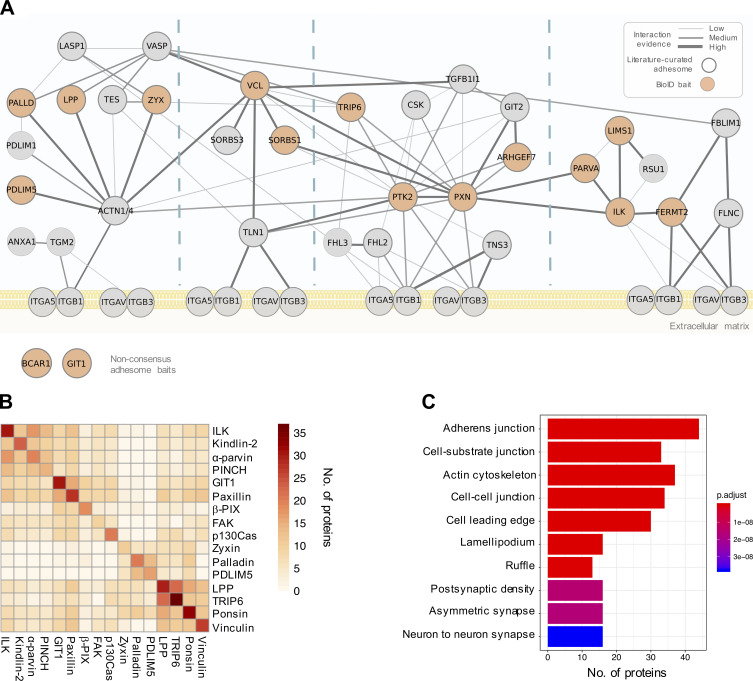
Figure 1.
Overview of the proximity-dependent adhesome. (A) 16 adhesome proteins were selected as BioID baits, 14 of which were present in the consensus adhesome and span the four putative signaling axes of the PPI network (Horton et al., 2015). p130Cas (Bcar1) and GIT1 are present in the literature-curated adhesome (and the GIT1 homologue GIT2 is in the consensus adhesome; Zaidel-Bar et al., 2007). Baits are shown in orange, and edges represent evidence of PPIs. Thick gray borders indicate literature-curated adhesome proteins. Gene names are shown. Consensus adhesome components unconnected to the main network are not shown. (B) Pairwise comparisons of proximal proteins (BFDR ≤ 0.05) identified by each BioID bait are displayed as a heatmap. Protein names not matching the gene names in A are FAK, PTK; kindlin-2, FERMT2; palladin, PALLD; α-parvin, PARVA; paxillin, PXN; β-Pix, ARHGEF7; PINCH, LIMS1; ponsin, SORBS1; vinculin, VCL; and zyxin, ZYX. (C) GO enrichment analysis of the 147 proteins in the proximity-dependent adhesome. The top 10 overrepresented terms under the cellular component category are shown.