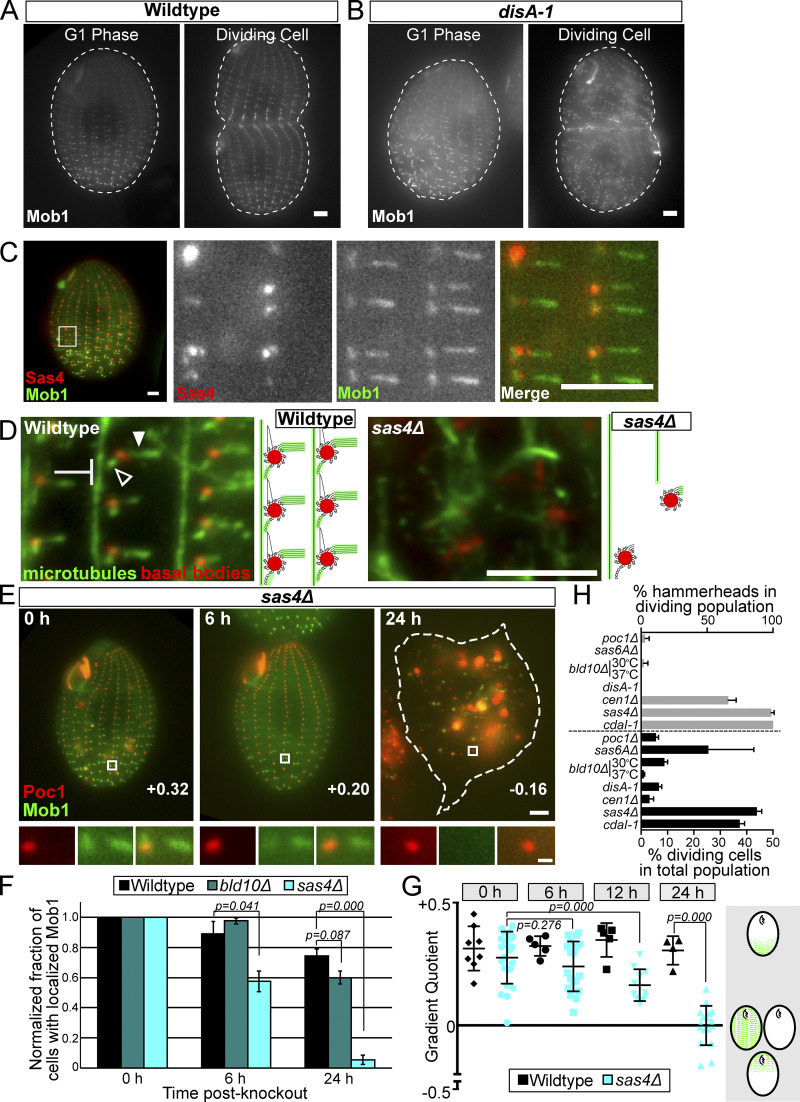
Figure 4.
Sas4 is required for Hippo factor localization to the cell cortex. (A) GFP:Mob1 is asymmetrically localized as a gradient from the posterior to anterior of WT G1 phase cells and its localization is enriched above the division furrow in dividing WT cells. Mob1 localizes to BBs and tMTs. Scale bar, 5 µm. (B) GFP:Mob1 localization in G1 phase and dividing cells in the cortical disorganization mutant, disA-1. Mob1 localizes as a posterior to anterior gradient at BBs and tMTs, as in WT cells. Scale bar, 5 µm. (C) Sas4:HaloTag (JF549) and GFP:Mob1 colocalize at daughter basal bodies. Mob1 localizes along the length of tMTs, but not at the proximal ends adjacent to BBs. Conversely, Sas4 localizes to the proximal end of tMTs that are not labeled with Mob1. Scale bars, 5 µm. (D) Cortical microtubules are lost and disorganized in sas4Δ cells. WT and sas4Δ cells at 24 h post-knockout labeled for BBs (red, centrin) and MTs (green, α-tubulin). Images focus on the cortical microtubules: tMTs (filled arrowhead), pcMTs (empty arrowhead), and longitudinal MTs (blunted arrow). Scale bar, 5 µm. (E) sas4Δ cells expressing GFP:Mob1 and Poc1:mCherry at 0, 6, and 24 h post-knockout. Scale bar, 5 µm. White boxes indicate the enlarged region below. Scale bar, 500 nm. Gradient quotient (determined in G) for each image is shown. (F) Quantification of the percentage of the cell population with proper Mob1 localization. Mob1 fails to localize in sas4Δ cells starting at 6 h post-knockout and is reduced to less than 10% of the cell population by 24 h after SAS4 knockout. At least 100 cells per condition and time point were quantified, and the experiment was performed in triplicate. Error bars denote SEM. Representative images of WT, bld10Δ, and sas4Δ cells at 24 h are shown in Fig. S3 I. (G) Quantification of Mob1 gradient in WT and sas4Δ cells at 0 h, 6 h, 12 h, and 24 h post-knockout. The gradient quotient (see Materials and methods) is calculated such that positive numbers indicate Mob1 enrichment in the posterior half of the cell and negative numbers indicate enrichment in the anterior half of the cell. A zero value indicates no gradient, because of either equally high or equally low amounts of Mob1 in both halves of the cell. Experiment was performed in duplicate, and each point on the graph represents a single cell. Error bars denote SD. (H) Quantification of cell division percentages and hammerhead cells in BB, cortical (disA-1), and Hippo (cdaI-1) mutants. Data shown are the average number of hammerheads seen and percent dividing cells during knockout or temperature shift of the indicated gene. At least 100 cells were counted per cell line per experiment. Experiment was performed in duplicate for poc1Δ, sas6AΔ, and cen1Δ, in triplicate for sas4Δ, bld10Δ at 30oC, and cdaI-1, and in quadruplicate for disA-1 and bld10Δ at 37oC. Error bars denote SEM.