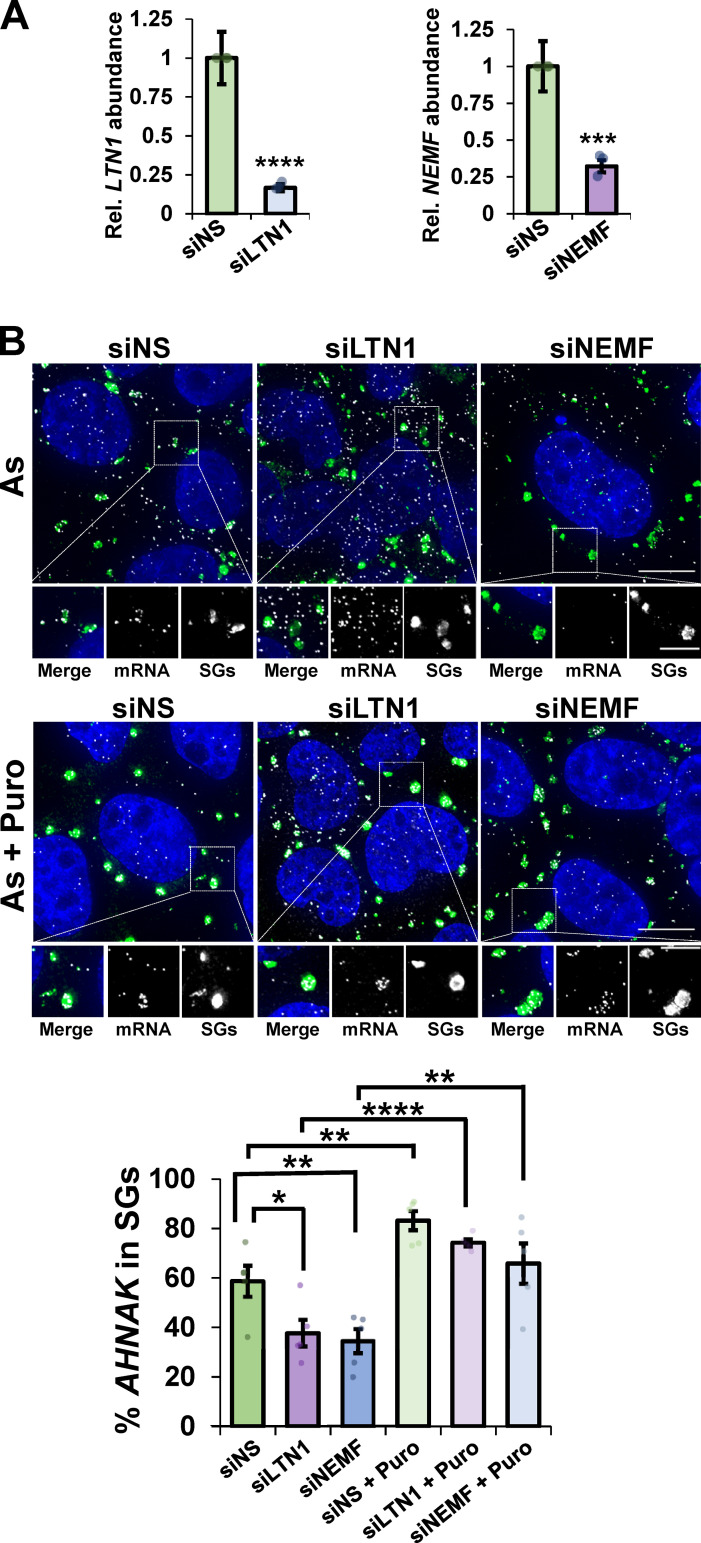
Figure 4.
Canonical RQC factors LTN1 and NEMF are required for efficient mRNA partitioning into SGs. U-2 OS cells were transfected with nonspecific siRNAs (siNS), siRNAs to LTN1 (siLTN1), or siRNAs to NEMF (siNEMF). (A) The levels of LTN1 and NEMF relative to GAPDH were detected by RT-qPCR (n = three independent experiments) with average ± SEM shown. Student’s t test was done to assess significance, with ***, P ≤ 0.005 and ****, P ≤ 0.001. (B) Cells were stressed for 45 min with arsenite (As; 0.5 mM) in the presence (As + puromycin [Puro], bottom panels) or absence (As, top panels) of Puro (10 µg/ml) with n = five frames counted in all conditions (42–57 total cells per condition). IMF staining to detect G3BP (green) and smFISH to detect AHNAK mRNA (white) was performed, and nuclei were visualized with DAPI (blue). Top: Representative photomicrographs. Scale bars, 10 µm (whole cell) or 5 µm (magnified panels). Bottom: The average percent AHNAK mRNA in SGs ± SEM is shown with individual points representing a single frame. Student’s t test was done to evaluate significance between siNS and siLTN1 or siNEMF conditions and between siRNA treatments or siRNA + Puro treatments, with *, P ≤ 0.05; **, P ≤ 0.01; and ****, P ≤ 0.001. Exact P values and source data are provided in Table S1.