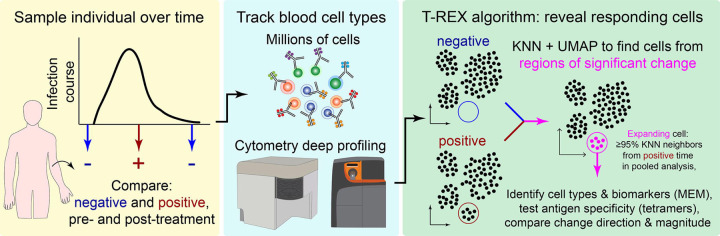
Figure 1 – Tracking Responders EXpanding (T-REX) algorithm identifies rare cells based on significant expansion or contraction during infection or treatment.
Graphic of the Tracking Responders Expanding (T-REX) workflow. Data from paired samples of blood from a subject are collected over the course of infection and analyzed by high dimensional, high cellularity cytometry approaches (e.g., Aurora or CyTOF instrument, as with datasets here). Cells from the sample pair are then equally subsampled for UMAP analysis. A KNN search is then performed within the UMAP manifold for every cell. For every cell, the percent change between the sample pairs is calculated for the cells within its KNN region. Regions of marked expansion or contraction during infection are then analyzed to identify cell types and key features using MEM. For some datasets, additional information not used in the analysis could be assessed to determine whether identified cells were virus-specific. Finally, the average direction and magnitude of change for cells in the sample was calculated as an overall summary of how the analyzed cells changed between samples.