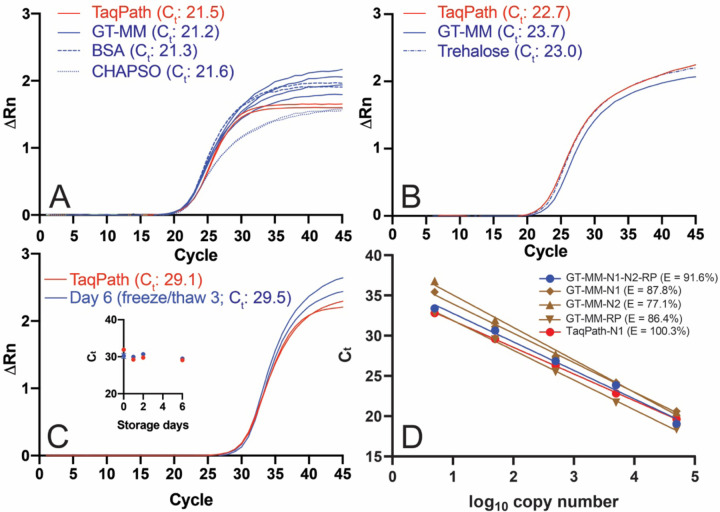
Figure 3. Performance of GT RT-qPCR master mix.
RT-qPCRs were performed with the Georgia Tech thermal cycling conditions (Table 2) and GT-Master Mix components (Table 4) with ATCC synthetic viral RNA (ATCC® VR-3276SD™) and Ct values determined using a threshold of 0.1 unless otherwise noted. (A) Effect of CHAPSO (0.1%) and BSA (0.5 mg/mL) on GT master mix performance with IDT N1 primers and 50,000 copies of synthetic viral RNA. The no template control did not amplify. (B) Performance of GT multiplex primers and probes with 50,000 copies viral RNA with GT-Master Mix, compared to TaqPath, and effect of trehalose (9.5%). (C) Performance of GT-Master Mix (MM) with IDT N1 primers and 500 copies of synthetic viral RNA, compared to TaqPath, after three freeze/thaw cycles (six days of storage) at 2x concentration. Inset: Ct for GT-Master Mix and TaqPath over six days of storage. (D) qPCR efficiency (E = 10 (−1/slope) −1) using auto threshold. GT-Master Mix and GT multiplex primers (N1 and N2 FAM readout, blue): 91.6%; GT-Master Mix and GT singleplex primers (brown): 87.8% for GT-N1 primer/probe (diamond), 77.1% GT-N2 primer/probe (triangle), 86.4% GT-RP primer/probe (inverted triangle); 100.3% TaqPath with GT-N1 primer/probe (red). Singleplex RT-qPCRs were performed with a mix of full-length viral RNA and HEK293T total RNA.