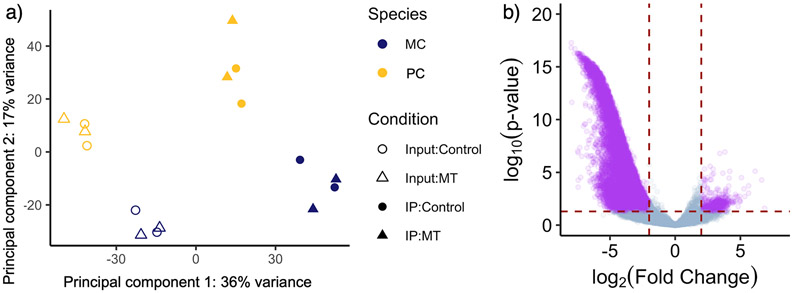
Figure 3:
a) Scatterplot of principal component 1 vs principal component 2 reveals clustering of overall expression profile by sample type and species, but not by condition. Principal component 1 accounts for 36% of all gene expression variation and, by inspection, separates the Input (open circles & triangles) and IP samples (closed circles and triangles). Principal component 2 accounts for 17% of the variation in gene expression and varies significantly with species. b) Volcano plot showing gene expression fold-difference and significance between input and IP samples (Model 1). We found that 1,443 genes are up-regulated in the IP samples while 14,786 genes are up-regulated in the input samples. Genes for which the adjusted p-value < 0.05 and where the Log2 Fold Change > 2 are shown in purple.