FIGURE 1.
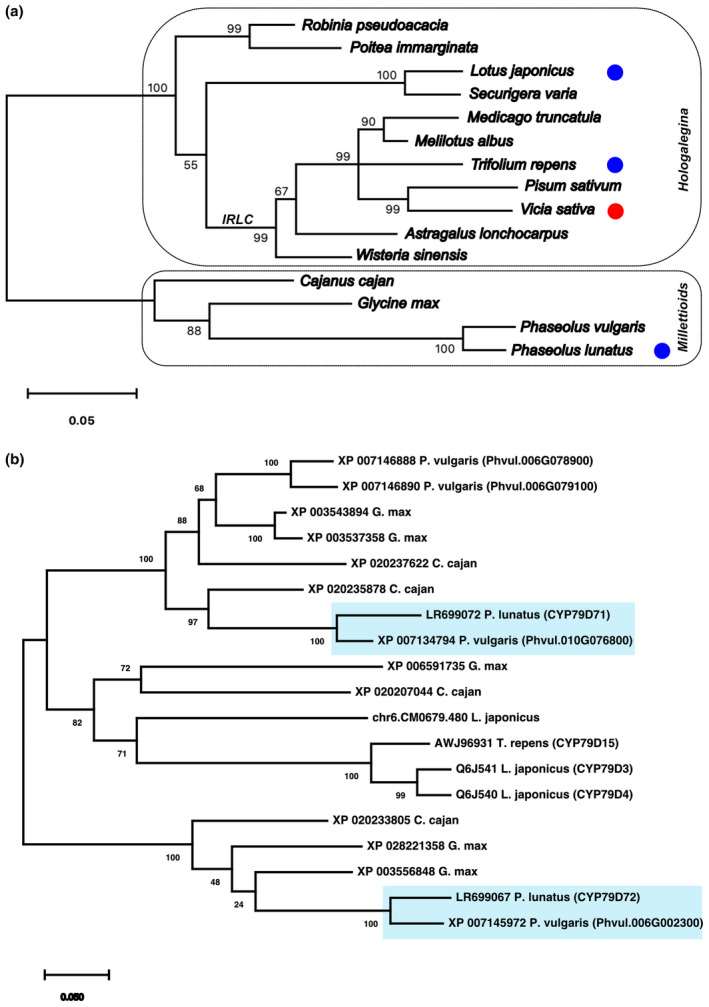
(a) Simplified phylogenetic tree of selected species belonging to the NPAAA‐clade within the legume family. It shows the Millettioid subclade containing the genus Phaseolus, and the Hologalegina subclade containing the genera Lotus, Trifolium, and Vicia. The presence of cyanogenic glucosides in a species is indicated by circles: linamarin/lotaustralin (blue) and prunasin/vicianin (red). The age of the Hologalegina diversification is estimated at about 50 million years ago, and that of the Millettioid clade at 45 million years ago (Lavin, Herendeen, & Wojciechowski, 2005). IRLC indicates the inverted‐repeat‐lacking clade. The phylogenetic analysis is based on chloroplast matK amino acid sequences. For a comprehensive overview of legume phylogeny see Wojciechowski et al. (2004). (b) A phylogenetic analysis of legume cytochrome P450 enzymes of the CYP79D‐subfamily. CYP79 protein sequences from six legume species are included: P. lunatus (Lima bean), P. vulgaris (common bean), Glycine max (soybean), Cajanus cajan (pigeon pea), Trifolium repens (white clover), and Lotus japonicus. Names represent GenBank accession numbers and/or with chromosomal locations and assigned names in parentheses. Phylogenetic analyses were performed with the Maximum Likelihood method and the Jones‐Taylor‐Thornton (JTT) matrix‐based model for amino acid sequences, using the MEGA X software. Positions containing gaps were eliminated and bootstrap values (1000x) are indicated at the branch points. Branch lengths are measured in the number of substitutions per site
