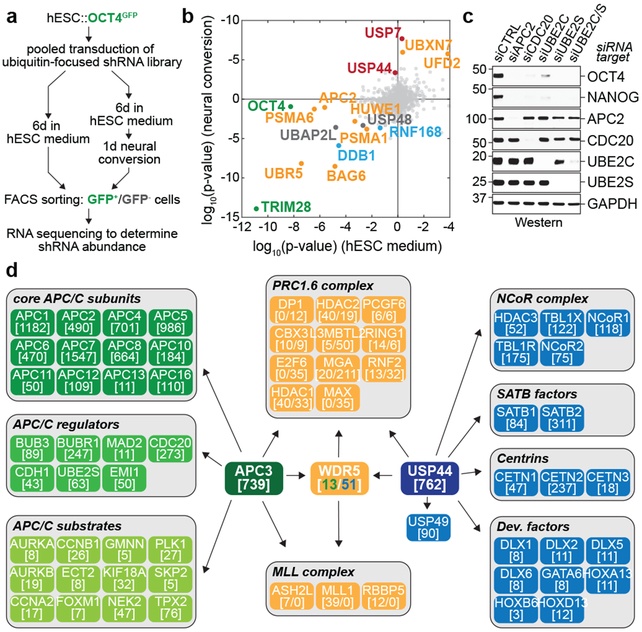
Fig 1. ∣. The APC/C stabilizes hESC identity.
a, Schematic of the ultracomplex shRNA screen.
b, shRNA screen identifies genes important for pluripotency. Each dot (n=886 unique genes) represents a gene’s p-value (Mann Whitney U test, two-sided, not corrected for multiple hypothesis testing) calculated from comparing the collection of shRNAs targeting each gene to all negative control shRNAs measured in each subpopulation (low versus high OCT4GFP levels). Orange: enzymes or effectors of K11/K48-branched chain synthesis; red: DUBs opposing K11/K48-specific E3s; blue: DNA repair enzymes; green: positive controls.
c, Western blot of pluripotency markers upon APC/C subunit knockdown in asynchronous H1 hESCs. This experiment was performed five independent times with similar results.
d, Interaction network of APC/C, WDR5, and USP44. Values listed in brackets are total spectral counts (TSCs) of tryptic peptides of indicated proteins.