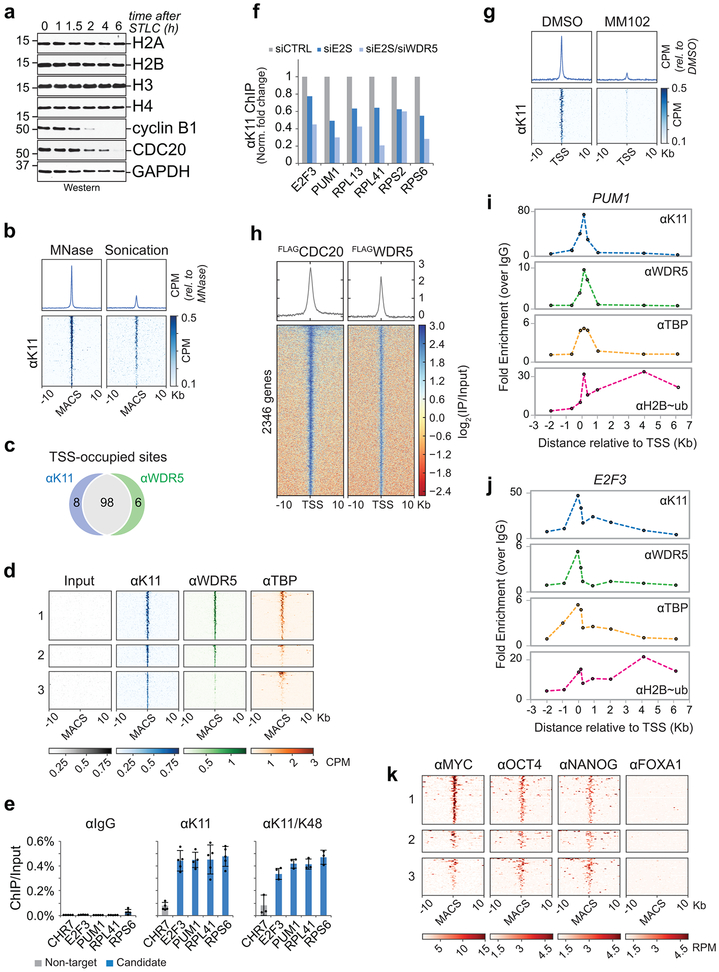
Extended Data Figure 8 ∣. ChIPseq analyses of APC/C- and WDR5-occupied gene targets.
a, Overall histone levels do not change upon release from mitosis. H1s were synchronized in mitosis by STLC and released into fresh medium. Indicated proteins were monitored by Western blotting. This experiment was performed two independent times with similar results.
b, Comparison between ChIPseq versus MNChIPseq against αK11 from mitotic H1 hESCs reveals that sonication shears polymeric ubiquitin linkages.
c, Venn diagram of αK11 and αWDR5 ChIP peaks that co-localize with TSSes from MNChIPseq experiments. MNChIPseq experiments were performed from mitotic H1 hESCs.
d, Heatmap of MNChIPseq data from mitotic H1 hESCs. Cluster 1 includes sites that are co-occupied by K11 and WDR5 near TSSes (within 100 bp), cluster 2 includes sites that are co-occupied by K11 and WDR5 outside of TSSes, and cluster 3 includes sites only occupied by K11, regardless of co-localization with TSSes.
e, ChIP-qPCR analysis of candidate targets using K11- or K11/K48-linkage specific ubiquitin antibodies from mitotic H1 hESCs (mean of independent replicates ± SD, n=3 for K11/K48, n=5 for IGG and K11 (except n=4 for PUM1)).
f, ChIP-qPCR analysis of mitotic H1 hESCs shows that K11-linkages synthesized at candidate sites are dependent on UBE2S and WDR5. This experiment was performed once.
g, WDR5 inhibition prevents K11-ubiquitin chain formation at APC/CWDR5-bound TSSes. H1 hESCs were treated with or without 50 μM MM102 during mitotic synchronization with STLC prior to αK11-MNChIPseq. Heatmap of all APC/CWDR5-bound TSSes are shown.
h, Heatmap of ChIPseq peaks of individual genes co-occupied by FLAGCDC20 and FLAGWDR5. ChIPseq against αFLAG was performed on mitotic HEK 293T cells overexpressing FLAGCDC20 or FLAGWDR5.
i, Spatial profile of PUM1 of factor occupancy by ChIP-qPCR. This experiment was performed once.
j, Spatial profile of E2F3 of factor occupancy by ChIP-qPCR. This experiment was performed once.
k, Heatmap of MNChIPseq data of transcription factor binding. Previously published MNChIPseq data was obtained from ref. 34, and APC/C-bound sites were similarly analyzed as described in d.