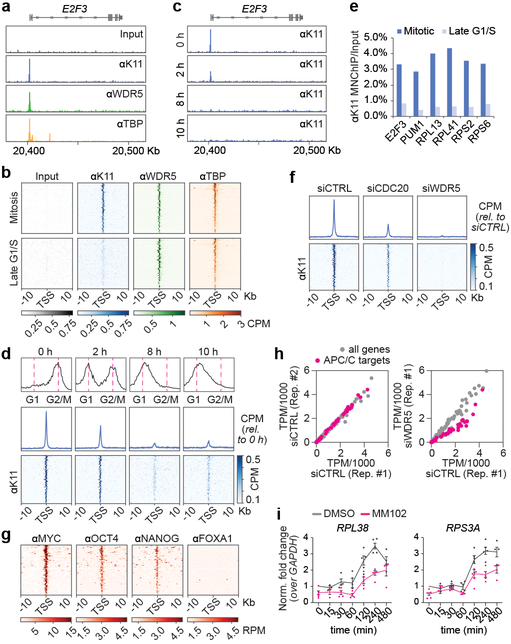
Fig 4. ∣. APC/C-dependent ubiquitylation occurs at TSSes of hESC genes.
a, Genome browser track of E2F3. MNChIPseq of indicated antibodies were performed from mitotic H1 hESCs.
b, K11 is deposited at select TSSes co-occupied by WDR5 in hESCs. Heatmap of co-occupied genes at TSSes from MNChIPseq experiments of indicated antibodies. H1 hESCs were collected after STLC treatment (mitosis) and after an 8h release (Late G1/S).
c, Genome browser track of E2F3 from MNChIPseq of αK11 in hESCs throughout a mitotic release.
d, Flow cytometry analysis of H1 hESCs upon mitotic synchronization and release into fresh medium (upper panels). Metagene analysis of K11- and WDR5-occupied TSSes (middle panels). Heatmap of individual K11- and WDR5-occupied TSSes from αK11-MNChIPseq experiments throughout a mitotic release (lower panels).
e, αK11-MNChIP-qPCR validates MNChIPseq findings that K11 is deposited only during mitosis in H1 hESCs. The same extract used in Fig. 4C was used for this experiment.
f, Depletion of CDC20 or WDR5 causes robust depletion of K11 chains at select TSSes.
g, MNChIPseq from HUES64 hESCs reveals that endogenous targets of APC/CWDR5 are strongly enriched in binding sites for MYC, OCT4, and NANOG.
h, Loss of APC/CWDR5 function interferes with expression of genes marked with K11-linked chains in H1 hESCs. Poly(A)-selected RNA was purified from asynchronous H1 hESCs transfected with siCTRL or siWDR5 for 48h and subjected to RNAseq.
i, RT-qPCR analysis of nascent RNA reveals APC/CWDR5 target genes are re-activated upon mitotic exit dependent on WDR5. Mitotic H1 hESCs were treated with or without 50μM MM102 and supplemented with 20μM zVAD-FMK. Cells were released into fresh media containing DMSO or 50μM MM102. RT-qPCR experiments were performed with oligonucleotides spanning intron-exon junctions. Values represent the mean of independent replicates ± SEM (n=3 for t=15 min, n=4 for t=30, 60, 480 min and n=5 for t=0, 120, 240 min).