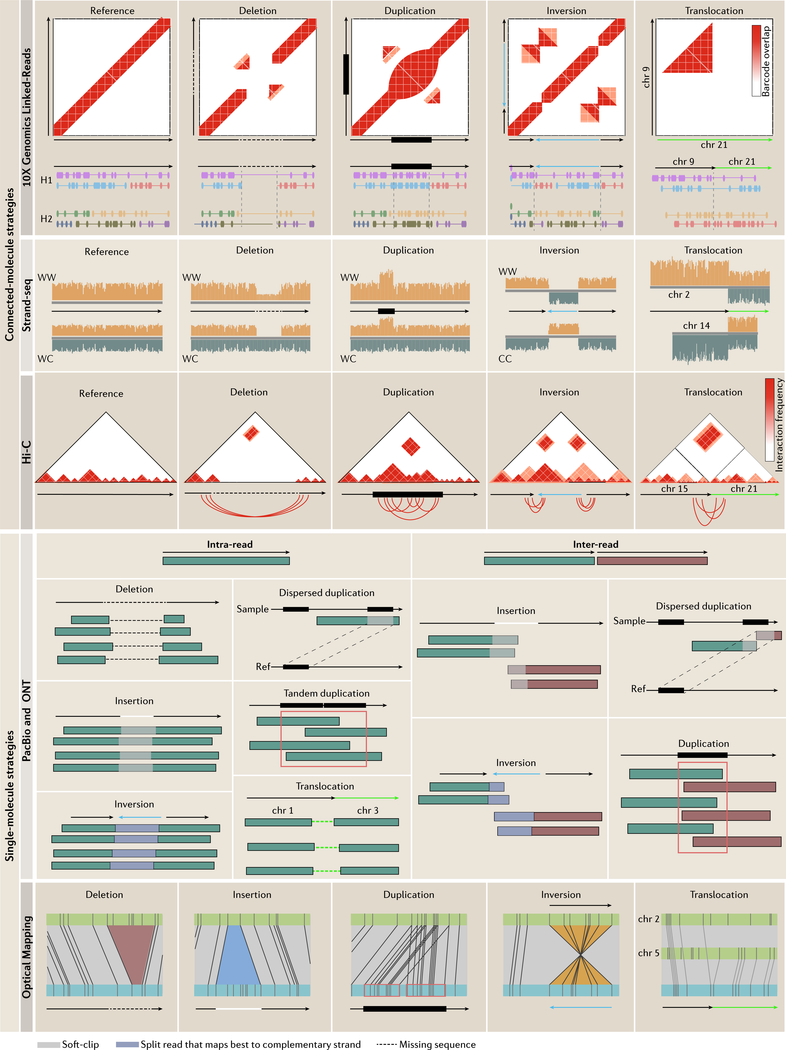
Figure 2 |. Structural variation signatures in single-molecule and connected-molecule strategies.
Emerging technologies vary in how they detect SVs. 10x Genomics linked-reads detect SVs based on barcode overlap between genomic loci. Split-molecule approaches infer SVs from splitting of linked-reads, examples of which are displayed below each barcode matrix (each color represents a shared barcode and linked-molecules are separated by haplotype; only homozygous variants are shown for simplicity). Strand-seq determines SVs based on read-depth or sudden changes in mapping orientation. For deletions and duplications, only two of four possible daughter cell configurations are shown for simplicity (Watson-Watson and Watson-Crick, Crick-Crick not shown). For inversions, only a homozygous inversion in Watson-Watson and Crick-Crick daughter cells are shown as Watson-Crick daughter cells mask homozygous inversions (homozygous for simplicity; for more detail on inversion detection see REF81. Hi-C detects SVs by looking for unusually high-frequency contacts between genomic loci. Underneath each interaction matrix is a schematic of the expected chromosomal contacts resulting from each SV. Single-molecule sequencing methods infer SVs based on discordant mapping signatures that can involve one (intra) or many (inter) reads. SVs derive from intra-read signatures, which result from reads that span an entire SV, or inter-read signatures, which require multiple reads to cover the event. Insertions differ from deletions by an increase in the expected distance between the two split pairs marked by the white soft-clip between the reads and inversions involve reads that map best to the complimentary strand. Optical maps detect SVs based on increased presence, absence or change in the orientation of restriction enzyme sites compared to a reference (blue: sample; green: reference). Resolution is dependent on the distribution of restriction enzyme sites.