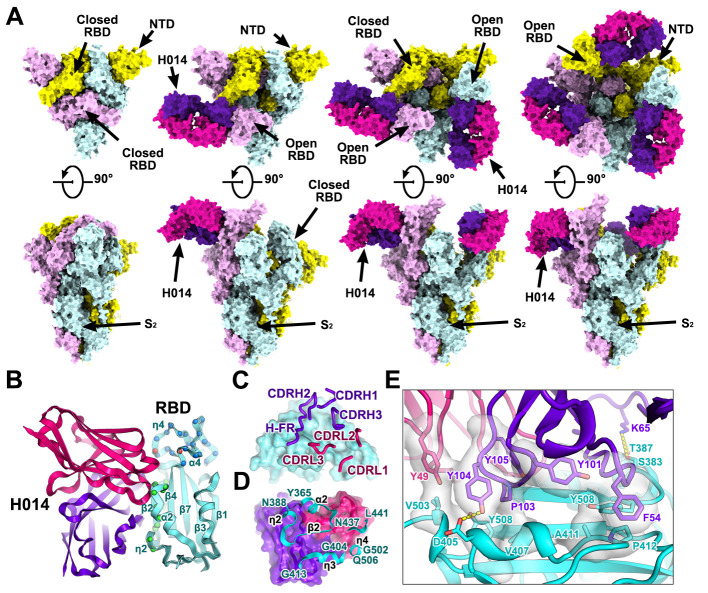
Fig. 2. Cryo-EM structures of the SARS-CoV-2 S trimer in complex with H014.
(A) Orthogonal views of SARS-CoV-2 S trimer with three RBDs in the closed state (left), one RBD in the open state and complexed with one H014 Fab (middle), two RBDs in the open state and each complexed with one H014 Fab. NTD: N-terminal domain. All structures are presented as molecular surfaces with different colors for each S monomer (cyan, violet and yellow), and the H014 Fab light (hotpink) and heavy (purpleblue) chains. (B) Cartoon representations of the structure of SARS-CoV-2 RBD in complex with H014 Fab with the same color scheme as in Fig. 2A. Residues comprising the H014 epitope and the RBM are shown as spheres and colored in green and blue, respectively. The overlapped residues between the H014 epitope and the RBM are shown in red. (C) and (D) Interactions between the H014 and SARS-CoV-2 RBD. The CDRs of the H014 that interact with SARS-CoV-2 RBD are displayed as thick tubes over the cyan surface of the RBD (C). The H014 epitope is shown as a cartoon representation over the surface of the RBD (D). (E) Details of the interactions between the H014 and SARS-CoV-2 RBD. Some residues involved in the formation of hydrophobic patches and hydrogen bonds are shown as sticks and labeled. Color scheme is the same as in Fig. 2A.