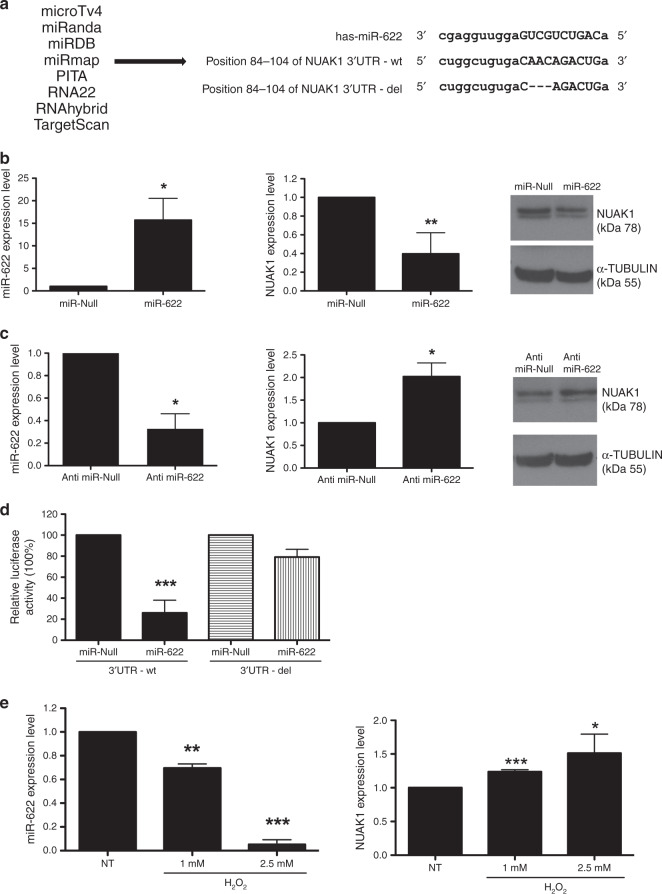
Fig. 2. NUAK1 is a direct target of miR-622.
a Shown is list of bioinformatics programs used to find the putative target of miR-622 (left) and the alignment of 3′UTR-NUAK1 wild type (−wt) and deleted (−del) version with the predicted binding site of miR-622 (right). b, c Expression levels of miR-622 (left) and NUAK1 (centre) mRNA were analysed by q-RT-PCR in MDA-MB-231 after transfection with miR-622 (b) or with Anti-miR-622 (c) respect to relative controls. β-ACTIN was used for normalisation. In b, c right, NUAK1 protein level was analysed in these cells by western blot, TUBULIN protein level was used as an endogenous control. d Luciferase activity was performed in MDA-MB-231/miR-622 and control cells co-transfected with NUAK1 3′UTR-wt or with 3′UTR-del. e MDA-MB-231 was treated with different doses of H2O2 and after 24 h, q-RT-PCR was performed to analyse miR-622 (left) and NUAK1 (right) expression levels. Each bar represents the mean ± SD of independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001.