Figure 2. CDC7 inhibition suppresses histone H2AX phosphorylation independently from origin firing.
-
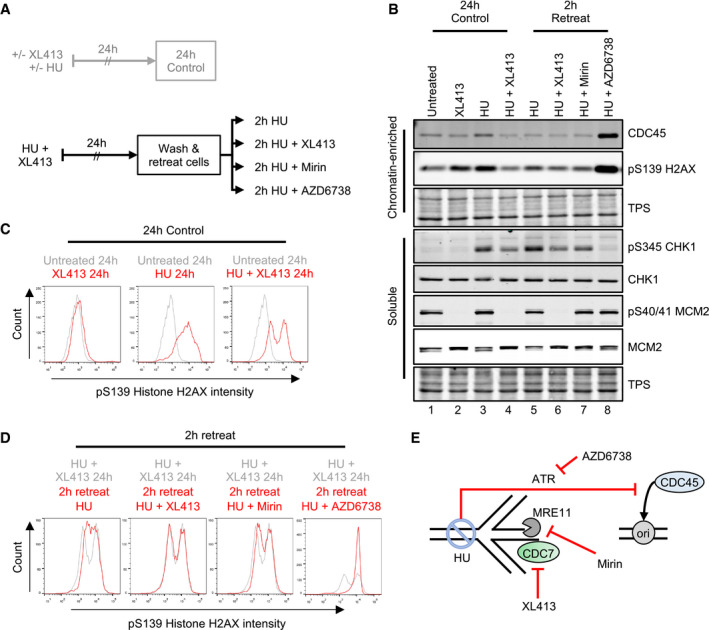
AOutline of experimental procedure. U2OS cells were either untreated or treated with 10 μM XL413 or with 4 mM HU in the presence or absence of XL413 for 24 h. Cells were EdU‐labelled for 30 min before harvesting. Cells that had been treated with both XL413 and HU for 24 h were washed with equilibrated media and retreated for a further 2 h. All further treatments included 4 mM HU, to prevent further DNA synthesis in either the absence or presence of CDC7 (XL413), MRE11 (Mirin) or ATR (AZD6738) inhibitors.
-
BCSK soluble and chromatin‐enriched protein fractions were analysed by Western blotting with the indicated antibodies. Data are representative of two independent experiments.
-
C, DFlow cytometry analysis to assess the levels of pS139 histone H2AX (c). Histograms show the mono‐parametric analysis of cell count against pS139 histone H2AX intensity. Histograms are overlaid to appreciate changes in pS139 histone H2AX intensity upon treatment (red lines) relative to appropriate experimental baseline controls (grey lines). Data are representative of two independent experiments.
-
EGraphical concept: cells treated with HU and XL413 for 24 h would have stalled forks, which exist in a stable state due to inhibition of CDC7 and with an active ATR‐dependent origin firing checkpoint. Upon washing and retreating of cells in the presence of HU, the late origin checkpoint is maintained, in an ATR‐dependent manner, and accumulation of DNA damage (pS139 H2AX) can be monitored independently from origin firing in the absence or presence of CDC7 (XL413) or MRE11 (Mirin) inhibitor. AZD6738 was used as a control to investigate molecular events occurring upon loss of checkpoint caused by both fork collapse and loss of the inhibition of origin firing. The increase in chromatin binding of CDC45 was used as a surrogate marker for origin (ori) activation, pS139 histone H2AX was used to monitor fork stability, pS345 CHK1 was used to monitor the ATR‐checkpoint signalling, and pS40/41 MCM2 was used to monitor CDC7 kinase activity.
Source data are available online for this figure.
