Figure 3. CDC7 is a replisome‐associated protein, and its inhibition affects the localization of MRE11 at replication factories.
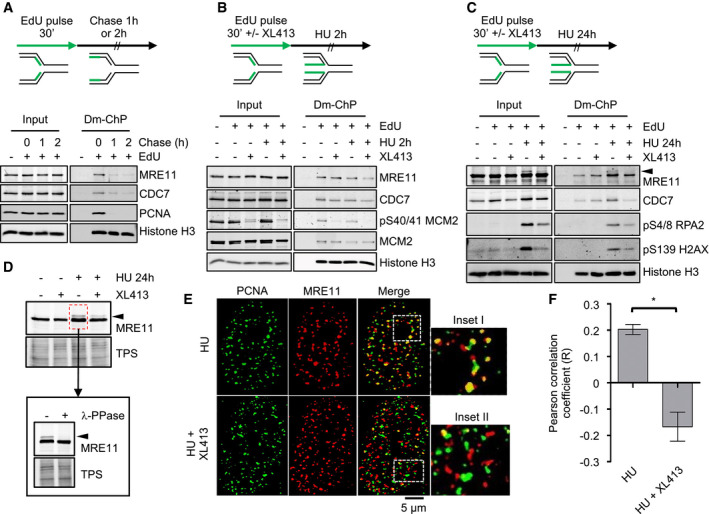
- U2OS cells were labelled with EdU for 30 min; then, EdU was washed, and cells were further incubated for either 1 or 2 h in the presence of thymidine. At the indicated time points, cells were fixed and proteins binding to EdU‐labelled DNA captured by the DNA‐mediated chromatin pull‐down technique (DmChP). Graphical experimental outline is shown above the analysis by Western blot of relevant proteins in both input and captured materials.
- U2OS cells were labelled with EdU for 30 min and then treated with 4 mM HU for 2 h in the presence or absence of 10 μM XL413. Proteins binding to EdU‐labelled DNA captured by DmChP were then analysed by Western blot as above.
- As in panel B, but incubation with HU was extended to 24 h. Black arrow indicates MRE11 electrophoretic mobility shift.
- U2OS cells were either mock‐treated or treated with 10 μM XL413 for 30 min, at which point 4 mM HU was added and cells incubated for a further 24 h. Extracts prepared from HU‐treated cells were then incubated in the presence or absence of λ‐phosphatase. Proteins were analysed by Western blotting with anti‐MRE11 antibodies. Total protein stain (TPS) is used as a loading control. Black arrow indicates MRE11 electrophoretic mobility shift.
- U2OS cells were treated with 4 mM HU in the presence or absence of 10 μM XL413 for 24 h. PCNA (green) and MRE11 (red) were detected by immunofluorescence. Insets I–II represent enlargements of selected region of the merged images.
- Quantification of PCNA and MRE11 colocalization was assessed with ImageJ in ˜70 randomly selected cells for each condition from four biological replicates and expressed as Pearson's correlation coefficient. Error bars represent SEM. Statistical significance was assessed by Student's t‐test (*P ˂ 0.05).
Source data are available online for this figure.
