-
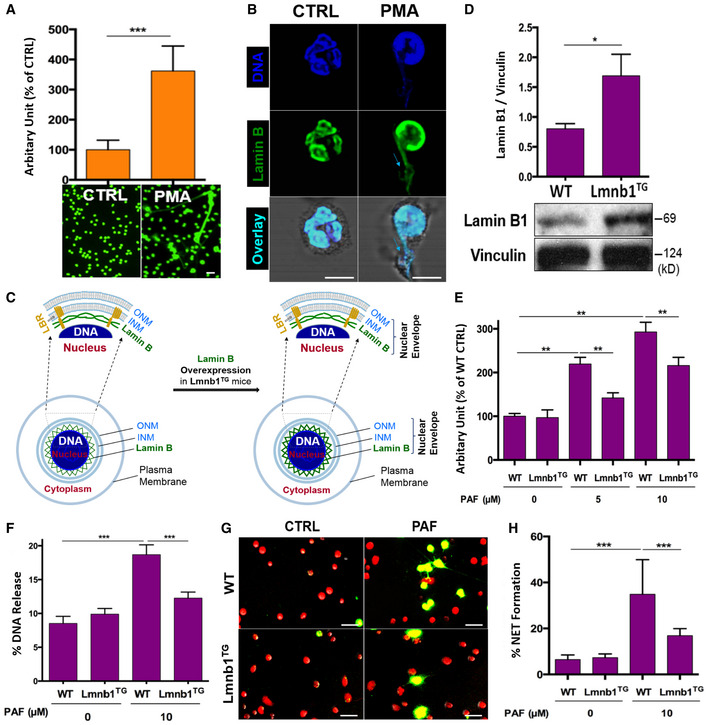
A
Summary and representative analysis of NET formation of pPMNs stimulated by 50 nM PMA for 3 h, detected by fluorescent microplate reader and confocal microscopy, respectively. Scale bar 20 μm.
-
B
Representative confocal microscopy images of pPMNs that were treated without (control) or with 50 nM PMA for 3 h and stained for lamin B and DNA as described in panel (B). The light blue arrows indicate the release of decondensed DNA associated with lamin B molecules from ruptured nuclear envelope. Scale bar 10 μm.
-
C
Schematic cross‐section of a cell and portion of the nucleus and the nuclear envelope, as well as the ones with corresponding overexpression of lamin B. The two lipid bilayers of the nuclear envelope are the inner and outer nuclear membranes (INM and ONM, respectively). The meshwork, nuclear lamin B is anchored to INM through lamin B receptor (LBR).
-
D
Representative and summary of immunoblots of lamin B of bone marrow neutrophils from WT and Lmnb1TG mice. Vinculin served as loading control.
-
E
Summary analysis of PAF‐induced NET formation in mPMNs from WT vs. Lmnb1TG mice that were stimulated without or with 10 μM PAF for 3 h and then fixed by 2% PFA and stained by SYTOX Green, followed by detection with fluorescent microplate reader.
-
F
The endpoint analysis of NET‐DNA release index was detected by coincubation of primary mouse peritoneal mPMNs from WT and Lmnb1TG mice without (control) or with 10 μM PAF in medium containing 1 μM SYTOX Green dye with recording by a microplate reader at the 3‐h time point. The NET‐DNA release index was reported in comparison with an assigned value of 100% for the total DNA released by neutrophils lysed by 0.5% (v/v) Triton X‐100.
-
G, H
Representative images (G) and summary analysis (H) of PAF‐induced NET formation in mPMNs from WT vs. Lmnb1TG mice that were stimulated without or with 10 μM PAF for 3 h and stained with both cell‐permeable SYTO Red and cell‐impermeable SYTOX Green, without fixation. Images were taken by Olympus confocal microscopy, followed by automated quantification of NETs on 5–6 non‐overlapping area per well using ImageJ for calculation of % cells with NET formation. (G) Scale bars, 40 μm.
Data information: Panels (A, D, E, F, H) were summary analysis of NET formation (A, E), or summary analyses of immunoblots (D), DNA% release index (F) that were calculated based on the arbitrary fluorescent readout unit (A, E), immunoblot arbitrary unit (D). NET‐DNA release index calculated based on fluorescent readout (F), or % cells with NET formation by image analysis (H). Data in (A, D, E, F, H) represent mean ± SD (
n = 3–5 biological replicates). *
P < 0.05, **
P < 0.01, ***
P < 0.001 between groups as indicated. Comparisons among three or more groups were performed using ANOVA, followed by Student–Newman–Keuls test. Comparison between two groups was analyzed by the Student
t test.
Source data are available online for this figure.