Figure 2.
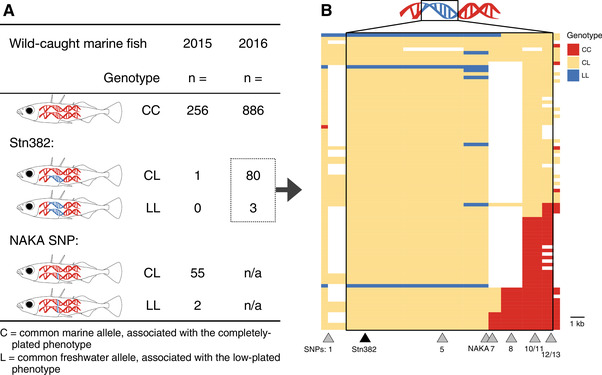
Freshwater alleles persist in the marine population. (A) Marine stickleback were collected and genotyped from Puget Sound in two consecutive summers, 2015 and 2016. In 2015, wild‐caught fish were sampled in midwater, genotyped first at the NAKA SNP, and subsequently at Stn382. The frequency of the L allele at the NAKA SNP was 9.4%, whereas the frequency of the full freshwater Eda haplotype was 0.16%. In 2016, fish were sampled nearshore, genotyped at Stn382, and the frequency of the L allele was 4.4%. (B) The carriers of the L allele at Stn382 from 2016 were genotyped at a subset of additional markers, and their genotypes are represented visually. Each row represents a single fish (n = 83), and the subset of markers genotyped within the haplotype (designated by the box) is shown below the plot. Triangles mark the physical location of SNP1 and the genotyped markers within the haplotype. Coloring representing the genotypes extends halfway to the next marker location. Additional markers on either side of the haplotype (Cnv767 and SNP19, ∼10 kb 5′ and 3′ of the region, respectively) were genotyped and are represented visually to the left of SNP1 and to the right of SNPs12/13, respectively. Missing data are in white.
