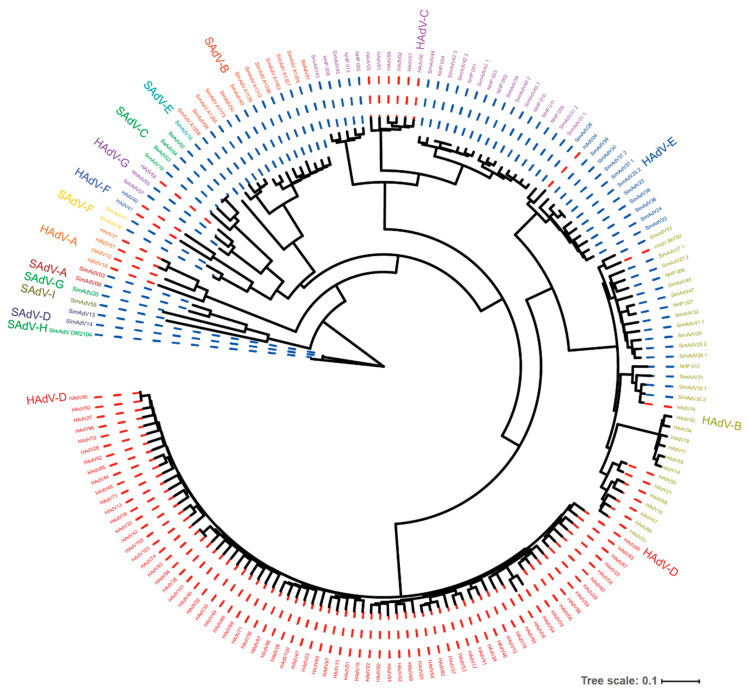
Figure 1.
Genetic resemblance of human- and non-human primate (NHP)-derived adenoviruses. A phylogenetic tree was constructed based on whole genome sequences of the adenoviruses that could be retrieved from Genbank. The Human and Simian Adenovirus species are color coded, and the colored dotted line indicates the adenovirus was isolated from either a human host (red) or NHP host (blue). The tree makes evident that the Human Adenovirus (HAdV) species HAdV-B, C, E, and G contain types isolated from human as well as from NHPs, suggesting cross-species transfer. The large group of HAdV-D types all originate from humans. The isolates numbered NHP-001 through NHP-013 represent new isolates from NHPs held in captivity (unpublished data). To generate the tree, sequences were assembled in FASTA format, aligned with MAFFT, and curated using BMGE. From the aligned data, a tree was generated using FastME. The process was performed at the NGPhylogeny.fr server by means of the “Oneclick workflow” using default parameters. The tree was exported to the Interactive Tree of Life server and visualized in circular mode. A high-resolution version of the figure is provided as Figure S1.