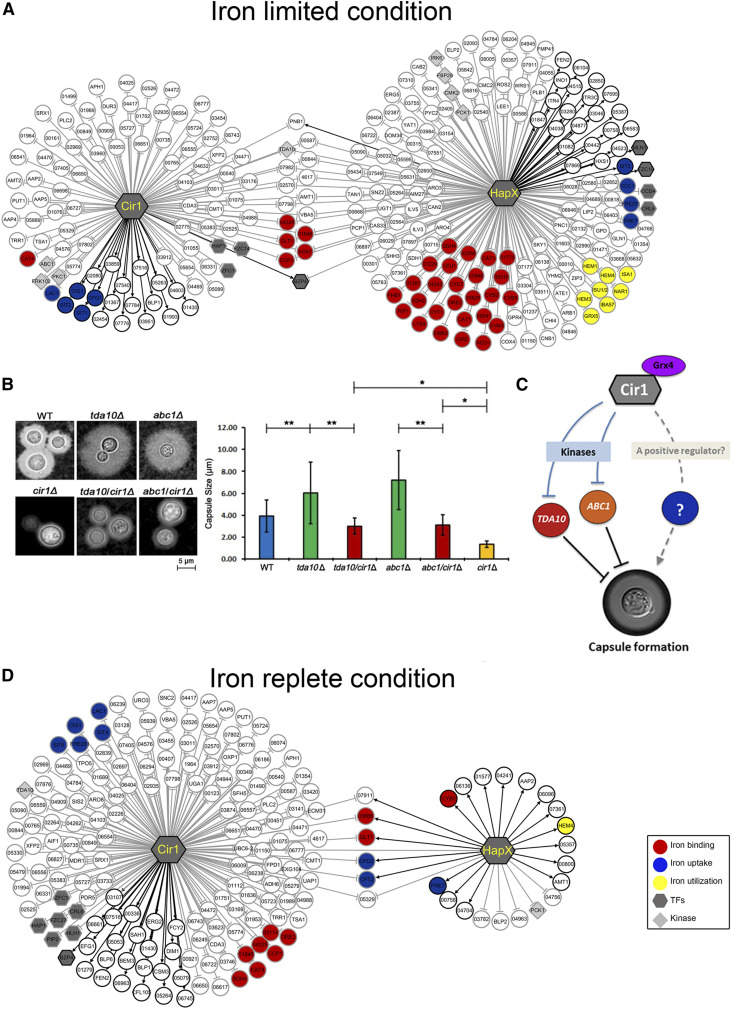
Figure 4.
The transcriptional networks of Cir1 and HapX. (A) Transcriptional regulatory networks are depicted to show the target genes of each transcription factor under iron-limited conditions. Genes regulated by Cir1 and HapX in an iron-independent manner are also included. (B) Capsules of C. neoformans strains grown in 1/10 sabouraud media were observed using a DCM310 microscope with India ink. Capsule sizes measured with 50 individual cells of each strain are shown in the graph. Note that the capsule size of the cir1Δ mutant is 1.36 ± 0.27 μm in these conditions. *P < 0.001; **P < 0.01. (C) Model for a possible regulatory mechanism of capsule formation based on direct regulation of Abc1 and Tda10 expression by Cir1. In the scenario shown, Cir1 indirectly regulates capsule formation by directly controlling expression of the genes encoding the protein kinases Tda10 and Abc1 which are negative regulators of capsule formation (Lee et al. 2016). A yet unknown regulatory factor, which might be a downstream of Cir1, may also play a positive regulatory role in capsule formation in C. neoformans. Additional regulatory scenarios are presented in the Discussion. (D) Transcriptional regulatory network depicting the target genes of each transcription factor under iron-replete conditions. Genes regulated by Cir1 and HapX in an iron-independent manner are also included. Black lines indicate positive regulation, and gray lines indicate negative regulation by the corresponding transcription factor.