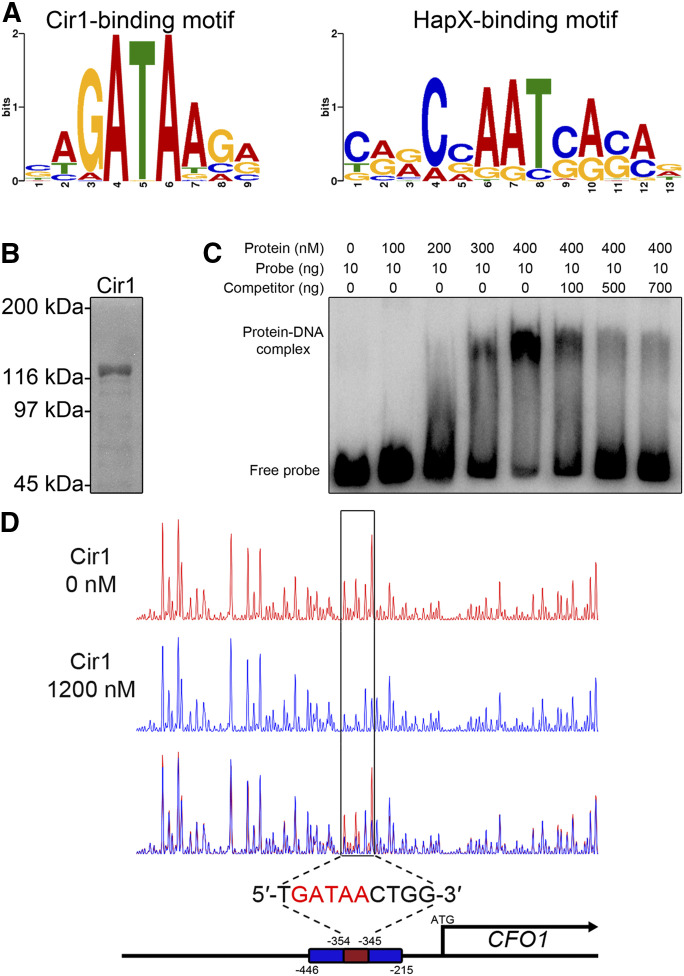
Figure 5.
Cir1 binds a GATA motif in the gene promoter region. (A) The binding motifs of Cir1 (left) and HapX (right) were identified using MEME analysis. (B) Full-length Cir1 protein was produced in the heterologous host E. coli, purified, and confirmed. (C) EMSA of DNA-protein binding was performed on a native polyacrylamide gel. The DNA probe was amplified from the CFO1 promoter region containing the predicted binding motif as a template. (D) DNase I footprinting using 6-FAM–labeled CFO1 promoter region was performed with (blue peaks) or without (red peaks) the purified Cir1 protein and analyzed by capillary electrophoresis. The position of the Cir1-binding motif was determined to span from −354 to −345 nt relative the transcription start site.