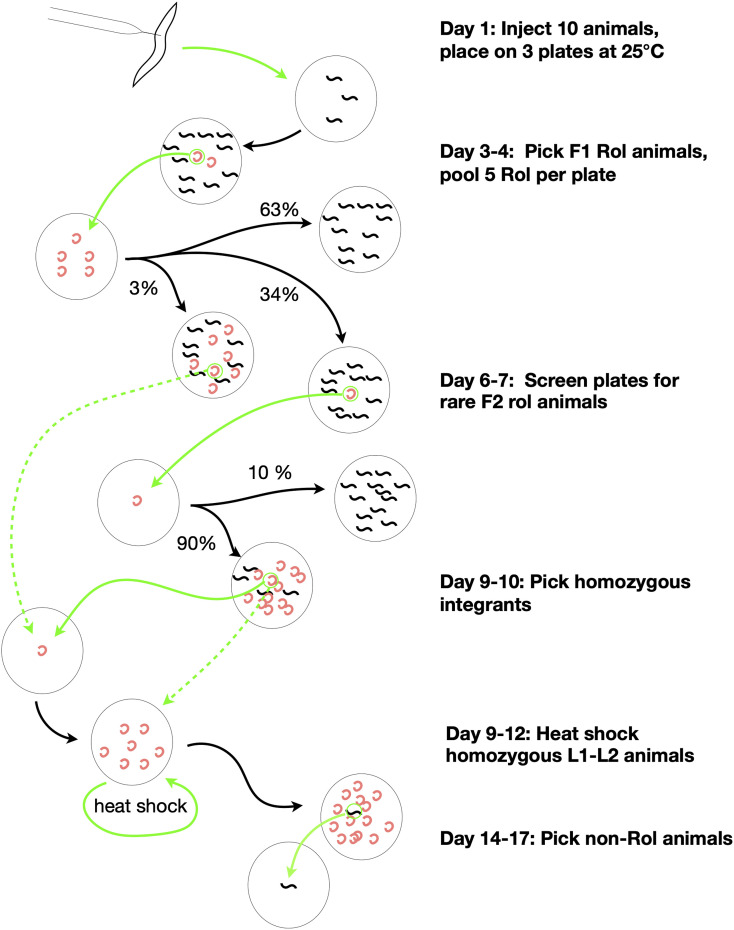
Figure 2.
Overview of RMCE procedure. A flow diagram illustrating the timeline of the RMCE procedure. The method consists of seven reliable steps. First, DNA of interest is inserted into pLF3FShC, a vector with a polylinker, which is compatible with SapI Golden Gate cloning (not shown). Second, miniprep DNA is injected at modest concentration (50 ng/µl) into the germline of animals carrying a landing site and injected animals are incubated at 25°. Third, on day 3, F1 Rol progeny are isolated and pooled ∼5 to a plate. Fourth, on day 6, “rare” F2 Rol are isolated as likely integrants. Fifth, on day 9, somatic non-GFP F3 homozygous animals are isolated. The loss of somatic GFP confirms the integration and provides a simple assay to identify homozygotes. Sixth, ∼20–30 L1–L2 homozygotes are heat shocked to induce Cre excision of the SEC. Finally, on day 14 or 15 non-Rol homozygous integrants are cloned. The large gray circles represent agar plates, small black squiggles represent wild type non-Rol worms and the red crescents represent Rol worms. Green arrows represent actions performed by the investigator and black lines represent one generation of growth of worms. Dashed arrows represent alternative time-saving steps that can occasionally be performed. The longer dashed arrow represents picking homozygous integrants on rare plates with early integration events. The smaller dashed arrow represents picking a large number of homozygous integrant individuals rather than allowing a single homozygous integrant to expand by selfing. The percentages represent the approximate fraction of plates which yield the distinct progeny distributions depicted.