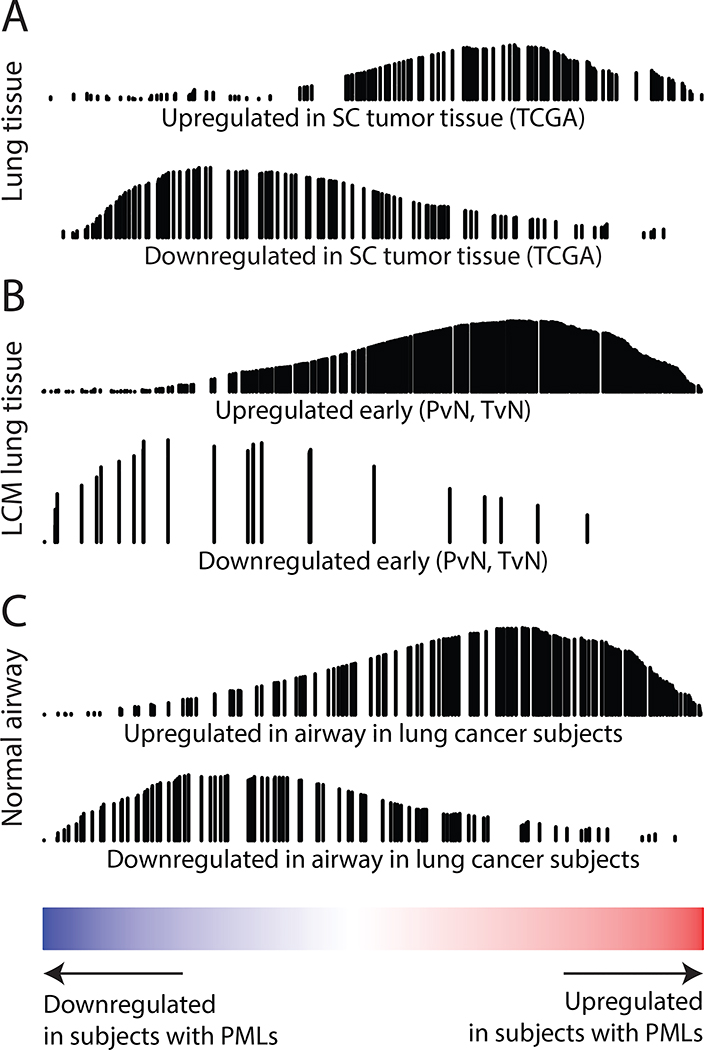
Figure 4. PML-associated gene expression alterations in the field are concordant with SCC-related datasets.
The genes up-regulated in the field of subjects with PMLs are red and genes down regulated in blue (x-axis). GSEA identified the significant enrichment of the lung cancer-related gene expression signatures shown in this ranked list. The black vertical lines represent the position of the genes in the gene set in the ranked list and the height corresponds to the magnitude of the running enrichment score from GSEA (y-axis). (A) Top differentially expressed genes from analysis of TCGA RNA-Seq data comparing lung SCC and matched adjacent normal tumor tissue. (B) Ooi et al. gene sets for early gene expression changes defined by genes altered between premalignant and normal tissue and between tumor and normal tissue (p<0.05) using laser capture microdissected (LCM) epithelium from the margins of resected SCC tumors. (C) Top differentially expressed genes from analysis of cytologically normal bronchial epithelial cells from smokers with and without lung cancer (GSE4115).