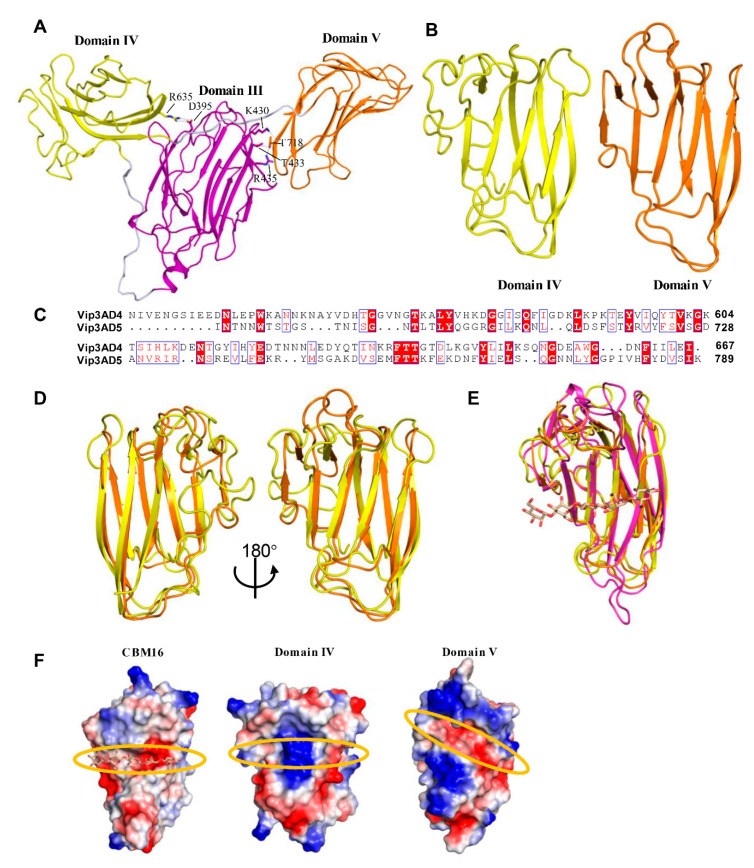
Figure 4.
Domains IV and V of Vip3Aa11 have glycan binding motifs. (A) Domain architectures of domains III, IV and V of Vip3Aa11. The polar interactions between domain IV, V and domain III are shown as sticks. (B) Overall structure of Vip3Aa11 domain IV and V shown as a ribbon cartoon. (C) Amino acid sequence alignment between domain IV and domain V of Vip3Aa11. The identical residues are denoted in white characters and red background, and the similar residues are denoted in red. ClustalX2 was used to perform the sequence alignment [35]. ESPript-3.0 was used to generate the figure [36]. (D) Two views of structure superimposition between domain IV and domain V of Vip3Aa11 shown as a ribbon cartoon. Color of each domain is consistent with Figure 4B. (E) Structure superimposition between domains IV, V of Vip3Aa11 and glycan bound CBM16 (RCSB ID 2ZEY) shown as a ribbon cartoon. Domains IV and V are colored as Figure 4B, and CBM16 is shown in magenta color. The glycan in CBM16 is shown as stick in light brown color. (F) Surface charge distribution of the sugar-binding pocket of CBM16 (RCSB ID 2ZEY) and potential sugar-binding pocket of domains IV, V of Vip3Aa11, highlighted with the orange circle.