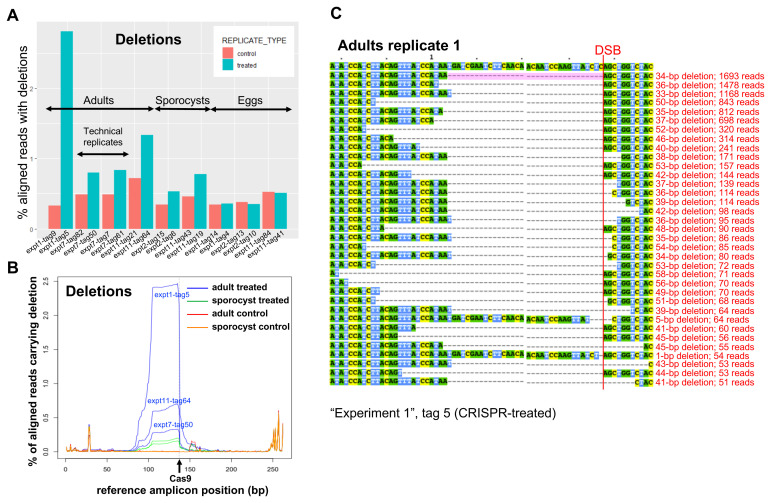
Figure 3.
( A) Frequency of deletions in NGS sequencing data, identified with the assistance of CRISPResso in three biological replicates from adults, two from sporocysts, and three from eggs, as indicated. ( B) CRISPR-induced deletions in adult worms and sporocysts. The positions of deletions found by CRISPResso in the reference amplicon are indicated, in three biological replicates of CRISPR-Cas9-treated adult samples (blue lines: experiment 1, tag 5; experiment 7, tag 50; experiment 11, tag 64) and matched adult control samples (red lines: experiment 1, tag 9; experiment 7, tag 82; experiment 11, tag 21), and in two biological replicates of CRISPR-Cas9-treated sporocysts (green lines: experiment 2, tag 6; experiment 11, tag 19) and matched sporocyst controls (orange lines: experiment 2, tag 15; experiment 11, tag 43). The black arrow shows the predicted Cas9 cut site. ( C) Multi-sequence alignment of SULT-OR alleles with deletions found in CRISPR-Cas9-treated adult worms that are supported by >=50 reads and span the DSB site indicated with a red line, based on one of the treated adult replicates (experiment 1, tag 5). The common 34-bp deletion is highlighted in pale pink.