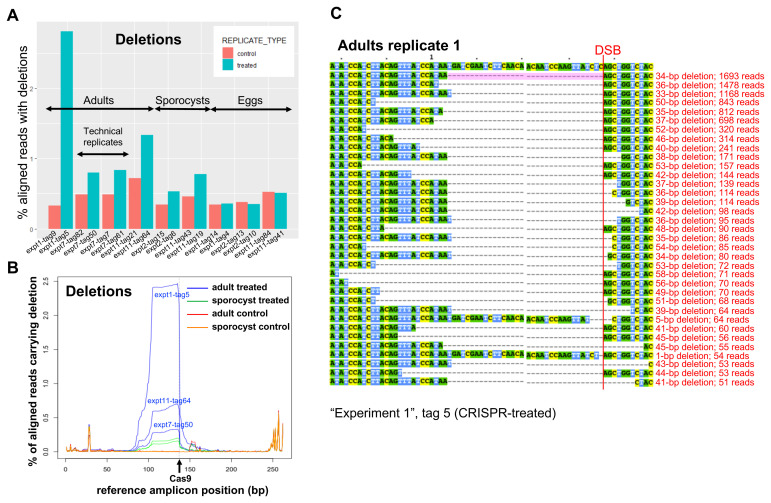
Figure 3.
( A) Frequency of deletions in NGS sequencing data, identified with the assistance of CRISPResso in three biological replicates from adults, two from sporocysts, and three from eggs, as indicated (sample identifiers at the bottom). The controls include worm-only controls for the adult samples, worm treated with Cas9-only and worm-only controls for the sporocyst samples, eggs treated with Cas9-only and egg-only controls for the egg samples. ( B) CRISPR-induced deletions in adult worms and sporocysts. The positions of deletions found by CRISPResso in the reference amplicon are indicated, in three biological replicates of CRISPR-Cas9-treated adult samples (blue lines: experiment 1, tag 5; experiment 7, tag 50; experiment 11, tag 64) and matched adult control samples (red lines: experiment 1, tag 9; experiment 7, tag 82; experiment 11, tag 21), and in two biological replicates of CRISPR-Cas9-treated sporocysts (green lines: experiment 2, tag 6; experiment 11, tag 19) and matched sporocyst controls (orange lines: experiment 2, tag 15; experiment 11, tag 43). The black arrow shows the predicted Cas9 cut site. ( C) Multi-sequence alignment of SULT-OR alleles with deletions found in CRISPR-Cas9-treated adult worms that are supported by >=50 reads and span the DSB site indicated with a red line, based on one of the treated adult replicates (experiment 1, tag 5). The common 34-bp deletion is highlighted in pale pink.