Abstract
As a hallmark of solid tumors, hypoxia promotes tumor growth, metastasis, and therapeutic resistance by regulating the expression of hypoxia-related genes. Hypoxia also represents a tumor-specific stimulus that has been exploited for the development of bioreductive prodrugs and advanced drug delivery systems. Cell division cycle 20 (CDC20) functions as an oncogene in tumorigenesis, and we demonstrated the significant upregulation of CDC20 mRNA in the tumor vs paratumor tissues of breast cancer patients and its positive correlation with tumor hypoxia. Herein, a hypoxia-responsive nanoparticle (HRNP) was developed by self-assembly of the 2-nitroimidazole-modified polypeptide and cationic lipid-like compound for delivery of siRNA to specifically target CDC20, a hypoxia-related protumorigenic gene, in breast cancer therapy. The delivery of siCDC20 by HRNPs sufficiently silenced the expression of CDC20 and exhibited potent antitumor efficacy. We expect that this strategy of targeting hypoxia-correlated protumorigenic genes by hypoxia-responsive RNAi nanoparticles may provide a promising approach in cancer therapy.
Keywords: hypoxia, polypeptide nanoparticle, RNA interference, gene silencing, CDC20, cancer therapy
Graphical Abstract

INTRODUCTION
The hypoxic microenvironment is common in the majority of solid tumors as a result of the imbalance between the increased oxygen consumption of the fast-growing tumor cells and the inadequate oxygen supply from the aberrant vasculatures,1,2 making it one of the most tumor-specific characteristics. Hypoxia is a critical regulator of tumor growth, malignant progression, and therapeutic resistance due to the hypoxia-mediated change of gene expression involved in angiogenesis, metabolism, cell proliferation, metastasis, and other cell processes.3-5 In this respect, up to 1.5% of the human genome is estimated to be correlated with hypoxia.6
With the remarkable reduction of oxygen in solid tumors compared to normal tissues, hypoxia also represents a unique stimulus for the development of tumor-specific diagnosis and therapy.7 Over the past decade, hypoxia-sensitive moieties, such as 2-nitroimidazole (NI) and azobenzene derivatives, were used as hypoxia markers (e.g., pimonidazole), radiosensitizers (e.g., misonidazole and etanidazole), and bioreductive prodrugs (e.g., tirapazamine and banoxantrone) for tumor hypoxia analysis and cancer treatment.8,9 More recently, several hypoxia-responsive nanoparticles (HRNPs) were developed for promoting controlled cargo release and enhancing cellular uptake for use in cancer therapy.10-15
The cell division cycle 20 (CDC20) gene plays a crucial role in mitotic progression by activating the anaphase-promoting complex/cyclosome (APC/C) E3 ubiquitin ligase to control the metaphase to anaphase transition.16 Moreover, CDC20 also regulates other cellular processes like apoptosis.17 It is well-documented that CDC20 functions as an oncogenic gene in tumorigenesis and is overexpressed in approximately 60% of breast cancers and many other human tumors.18 Knocking down CDC20 in various cancer cells has also been shown to result in a mitotic arrest followed by cell death.19 Although several small molecule inhibitors have been explored for CDC20 activity, their low specificity and/or blocking efficiency to CDC20 may hinder clinical translation and lead to severe side effects,20,21 highlighting the need for advanced strategies for more effective inhibition of this therapeutic target.
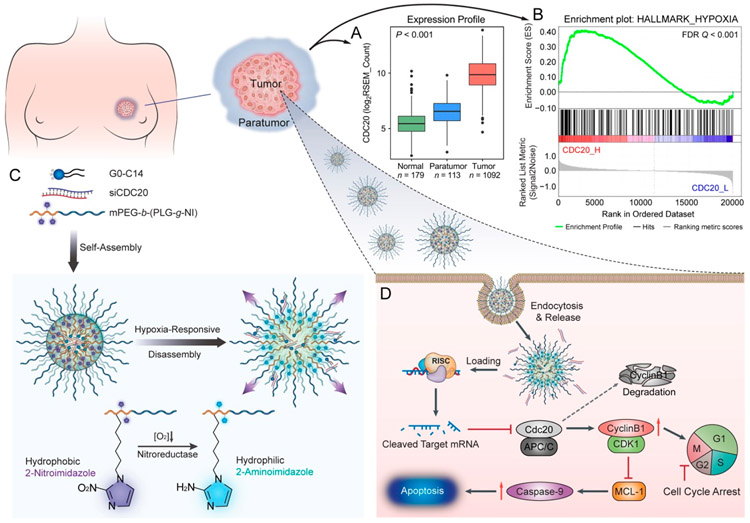
In this work, our bioinformatics analyses demonstrated that a strong correlation exists between high CDC20 mRNA expression and tumor hypoxia in breast cancer, and an increased level of CDC20 expression was also observed in tumor cells under hypoxic vs normoxic conditions (Figure 1A, B). We thus hypothesized that specific targeting of the hypoxia-correlated pro-tumorigenic gene CDC20 by hypoxia-responsive therapeutics would lead to the effective and safe treatment of breast cancer. To do this, we developed an HRNP platform composed of NI-modified polypeptide and a cationic lipid-like compound for the delivery of siRNA to silence CDC20 expression in breast cancer cells specifically, as shown in Figure 1C. Under the hypoxic tumor environment, conversion of the hydrophobic NI group to a hydrophilic 2-aminoimidazole (AI) occurs as a result of a series of nitroreductases that catalyze a single-electron reduction, leading to the disassembly of HRNP and a subsequent rapid release of cargo in tumor cells (Figure 1D). The small HRNP/ siRNA with a diameter of 54.7 nm also demonstrated high tumor accumulation, efficient CDC20 silencing in tumor tissue, and efficient tumor suppression in vivo, indicating the enormous potential of this dual hypoxia-targeting RNAi nanoparticle for breast cancer therapy.
Figure 1.
Hypoxia-responsive nanoparticle (HRNP) for targeting hypoxia-correlated protumorigenic gene (CDC20) in precision breast cancer therapy. (A) Significant upregulation of CDC20 mRNA was observed in breast cancer tissues compared to normal and paratumor tissues. (B) The GSEA result shows the positive correlation of CDC20 with tumor hypoxia. (C) Formulation of HRNP/siRNA and mechanism of disassembly in a hypoxic reduction environment. (D) Schematic of HRNP/siCDC20 delivery into tumor cell cytoplasm to induce G2/M arrest and cell apoptosis for effective cancer therapy.
RESULT AND DISCUSSION
Bioinformatics Analyses of CDC20 in Breast Cancer Samples and Cell Lines.
In the first set of experiments, we collected the RSEM-normalized RNA-Seq data of CDC20 from the Genotype-Tissue Expression (GTEx) data set and the Cancer Genome Atlas (TCGA) breast cancer data set. Figure 1A demonstrates that CDC20 mRNA was significantly elevated in 1092 tumor tissues compared to 179 donated normal tissues and 113 paratumor tissues. In 112 paired TCGA samples, the upregulation of CDC20 was observed in 110 (98.2% of 112) primary breast cancer tissues compared to corresponding paratumor tissues (Figure S1, Supporting Information). We also performed the Gene Set Enrichment Analysis (GSEA) to investigate the relationship between CDC20 expression and hypoxia by using the Cancer Cell Line Encyclopedia (CCLE) database (Figure 1B).22,23 The result of GSEA indicated that CDC20 expression was positively correlated with hypoxic status in breast cancer. Moreover, breast cancer patients with higher expression of CDC20 exhibited worse overall survival (OS) and relapse-free survival (RFS) (Figure S2, Supporting Information), indicating that the overexpression of CDC20 may contribute to therapeutic resistance in breast cancer patients, which is consistent with a previously reported study.24
Synthesis and Characterizations of Hypoxia-Responsive Polymer and HRNP/siRNA.
The hypoxia-responsive polypeptide, methoxy poly(ethylene glycol)-block-poly(L-glutamide-graft-2-nitroimidazole) (mPEG-b-(PLG-g-NI)), was synthesized by the condensation reaction between the carboxyl group in the LG unit and the amino group in 6-(2-nitroimidazole)hexylamine (NIHA). The successful synthesis of mPEG-b-(PLG-g-NI) was confirmed by the characteristic signals in the proton nuclear magnetic resonance (1H NMR) spectrum (Figure S5, Supporting Information). The numberaverage molecular weight (Mn) of mPEG-b-(PLG-g-NI) was 13 400 g mol−1, and the polydispersity index (PDI) was determined to be 1.58 by gel permeation chromatography (GPC; Figure S6, Supporting Information).
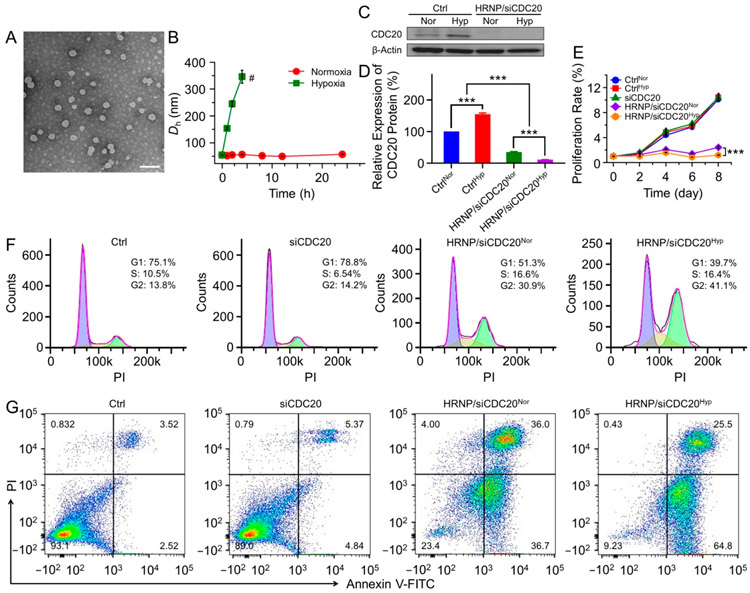
After successful synthesis of mPEG-b-P(LG-g-NI), HRNP was formulated by nanoprecipitation for siRNA delivery, as shown in Figure 1C. Briefly, the solution of mPEG-b-P(LG-g-NI) and cationic lipid-like compound G0-C14 in N,N-dimethylformamide (DMF) was mixed with a siRNA aqueous solution to form the siRNA-loaded HRNP.25 As shown in Figure 2A, the spherical morphology of HRNP/siRNA was revealed by transmission electron microscopy (TEM). The average hydrodynamic diameter (Dh) of HRNP/siRNA was about 54.7 nm with a PDI of 0.168, as measured by dynamic light scattering (DLS; Figure S7, Supporting Information). To confirm the hypoxia-responsiveness of HRNP/siRNA, the changes in size vs time were monitored. As depicted in Figure 2B, the size of HRNP remained nearly constant under normoxic conditions, while the size increased remarkably under hypoxic conditions. The resultant increase in size represents the conversion of the hydrophobic NI group to a hydrophilic AI and hypoxia-triggered disassembly of HRNP. This is consistent with previous reports11,12 and indicates the hypoxia-responsive capability of the nanoplatform.
Figure 2.
Characterizations of HRNP/siRNA and CDC20 gene silencing on MCF-7 cells in vitro. (A) TEM image of HRNP/siRNA. The scale bar is 100 nm. (B) Dh changes of HRNP/siRNA under normoxic vs hypoxic conditions in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (20.0 mM) at pH 7.4. (C) Western blot and (D) quantitative analyses of CDC20 expression in MCF-7 cells treated with HRNP/ siCDC20 under normoxic vs hypoxic conditions. (E) Proliferation profile, (F) cell cycle analyses, and (G) apoptosis analyses of MCF-7 cells treated with free siCDC20 or HRNP/siCDC20 under normoxic vs hypoxic conditions. The cells incubated in culture medium without HRNP or free siRNA were regarded as a control group. The statistical data are presented as mean ± SD (standard deviation; n = 3; ***P < 0.001).
In Vitro Gene Silencing.
To investigate whether the disassembly of HRNP/siRNA triggered by hypoxic conditions can improve gene silencing, the luciferase-expressing HeLa (Luc-HeLa) cells were first chosen as a model cell line.26 As shown in Figure S8, Supporting Information, HRNP/siLuc significantly suppressed the luciferase expression in Luc-HeLa cells when incubated in either normoxic or hypoxic conditions. More impressively, in hypoxic conditions, an even better gene silencing efficiency was achieved as approximately 90% of the luciferase expression was knocked down when treated with HRNP/siLuc at a siRNA dose of 10.0 nM, and no apparent cytotoxicity was observed. We then examined the CDC20 silencing by HRNP/siCDC20 in vitro using Western blot (Figure 2C,D). Similarly, the results showed that HRNP/ siCDC20 exhibited a better gene silencing effect under hypoxic conditions. Notably, the upregulation of CDC20 expression was observed under hypoxic conditions, which is consistent with the GSEA result (Figure 1B).
To understand the contribution of hypoxic conditions in enhancing the gene silencing, MCF-7 cells were treated with HRNP/Cy5-siRNA for 4 h under normoxic vs hypoxic conditions followed by an investigation of the cellular uptake and endosomal escape using flow cytometry (FCM) and confocal laser scanning microscopy (CLSM), respectively. Similar cellular uptake was observed under both normoxic and hypoxic conditions (Figure S9, Supporting Information), with slightly higher uptake in the hypoxic group. The less colocalization of Cy5-siRNA and endosomes, as indicated by the yellow area (Figure S10, Supporting Information), suggests that the hypoxic conditions could improve the endosome escape of siRNA nanoparticles. More efforts will be needed to clarify the underlying mechanisms further.
Next, we examined the effect of CDC20 silencing on MCF-7 cell proliferation. As shown in Figure 2E, significant inhibition of cell proliferation by HRNP/siCDC20 was detected in MCF-7 cells compared to the untreated group and free siCDC20 group. Additionally, the inhibitory effect was more profound when cells were incubated under hypoxic conditions. Previous studies have reported that CDC20 could regulate mitotic progression and apoptosis,19 and thus, we performed the cell cycle and apoptosis analyses on MCF-7 cells using FCM to investigate the mechanisms of inhibition on cell proliferation induced by HRNP/siCDC20. As shown in Figure 2F, HRNP/ siCDC20 increased the proportion of cells in G2 phase, especially under hypoxia, which is consistent with our GSEA result (Figure S11, Supporting Information). Furthermore, HRNP/siCDC20 also increased the apoptosis rate (Figure 2G) under hypoxic conditions. These results suggest that the reduced proliferation of MCF-7 cells upon CDC20 silencing was due to the increased proportion of cells in G2 phase and increased apoptosis rate, with hypoxic conditions further facilitating G2/M arrest and apoptosis.
Pharmacokinetics and Biodistribution.
To explore in vivo performance of the HRNP/siRNA, healthy and MCF-7 tumor-bearing female BALB/c nude mice were used for pharmacokinetic and biodistribution assays, respectively.27 The pharmacokinetic study was performed by the administration of free DY677-siRNA vs HRNP/DY677-siRNA (1.0 nmol of siRNA per mouse) through tail vein injection. Figure 3A shows that free DY677-siRNA had a very short circulation time (around 30 min), thus quickly disappearing from the blood. In contrast, HRNP/DY677-siRNA had a much longer blood circulation time due to the PEGylated surface, with around 10% of the initially injected siRNA nanoparticles still in circulation after 12 h. In the biodistribution study, free DY677-siRNA or HRNP/DY677-siRNA was iv injected into MCF-7 tumor-bearing mice, followed by harvesting of the major organs and tumor 24 h postinjection. HRNP/DY677-siRNA showed high accumulation in the tumor site at 24 h (Figure 3B,C), with a 6-fold higher fluorescence signal than that of free DY677-siRNA.
Figure 3.
In vivo pharmacokinetics and biodistribution of HRNP/siRNA. (A) Pharmacokinetic profiles, (B) biodistribution, and (C) quantification in major organs of free DY677-siRNA and HRNP/DY677-siRNA after iv injection. The dose of siRNA was fixed at 1.0 nmol per mouse. The statistical data are presented as mean ± SD (n = 3; ***P < 0.001).
In Vivo Gene Silencing and Antitumor Efficacy.
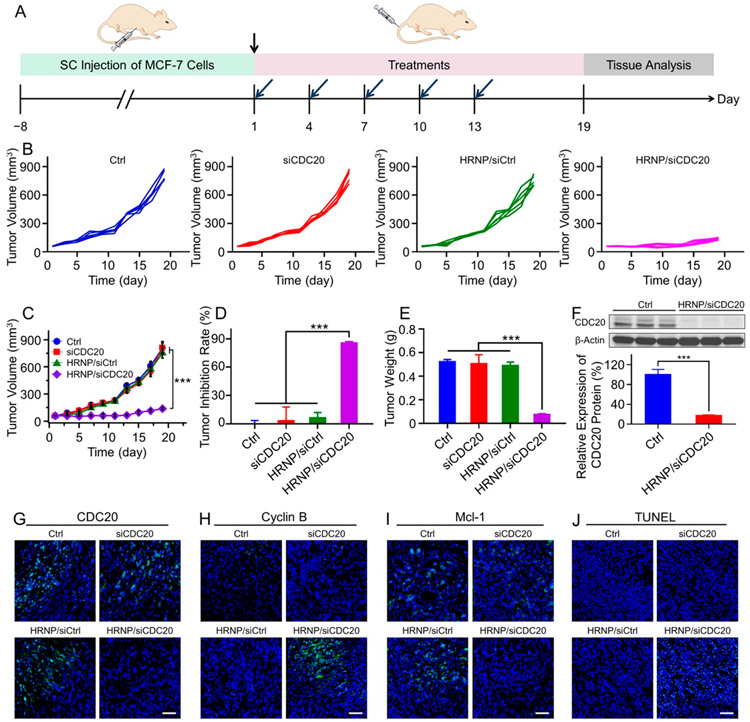
In the last set of experiments, we tested the in vivo antitumor efficacy of HRNP/siCDC20. First, MCF-7 tumor-bearing BALB/c nude mouse models were established and then administered PBS, siCDC20, HRNP/siCtrl, or HRNP/siCDC20 on day 1, 4, 7, 10 and 13 as shown in Figure 4A, and the dose of siRNA was fixed at 1.0 nmol/mouse. In regard to the xenograft breast tumor model with an initial tumor volume of 55 mm3, HRNP/ siCDC20 significantly inhibited the tumor growth (Figure 4B,C), and the antitumor efficiency was up to 85.6% (Figure 4D), when compared to PBS, siCDC20, and HRNP/siCtrl, without noticeable body weight changes during the treatment (Figure S12, Supporting Information). We also investigated the CDC20 and its downstream gene expression levels in the tumor tissues using Western blot and immunofluorescence staining. More than 90% of CDC20 knockdown at the protein level was observed in the HRNP/siCDC20 group (Figure 4F,G, and Figure S13, Supporting Information). The silencing of CDC20 expression upregulated cyclin B expression (Figure 4H and Figure S14, Supporting Information) and downregulated Mcl-1 expression (Figure 4I and Figure S15, Supporting Information), indicating that the significant tumor growth inhibition could be attributed to cell cycle arrest and apoptosis. Moreover, the terminal deoxyribonucleotidyl transferase (TDT)-mediated dUTP-digoxigenin nick end labeling (TUNEL) analyses of the tumor tissues showed high levels of apoptosis after the treatment of HRNP/siCDC20 as compared to PBS, siCDC20, and HRNP/siCtrl (Figure 4J and Figure S16, Supporting Information). Additionally, no obvious toxicity and inflammatory responses were observed in major organs from hematoxylin and eosin (H&E) staining (Figure S17, Supporting Information), and the hematological parameters, such as alanine aminotransferase (ALT) and blood urea nitrogen (BUN), were in the normal ranges (Figure S18, Supporting Information). Similarly, the antitumor efficacy of HRNP/siCDC20 was demonstrated in the MCF-7 tumor-bearing BALB/c nude mouse model with a higher initial tumor volume at about 120 mm3. As shown in Figure S19, Supporting Information, the tumor inhibition curves and tumor inhibition rates showed a similar trend to those from the mice with a smaller tumor volume in Figure 4, without noticeable body weight changes during the treatment (Figure S20, Supporting Information), demonstrating that HRNP/siCDC20 may serve as a potent nanoplatform for breast cancer therapy at various stages.
Figure 4.
In vivo antitumor efficacy and gene silencing of HRNP/siCDC20 on MCF-7 tumor-bearing BALB/c nude mice. (A) Timetable of construction of MCF-7 tumor-bearing BALB/c nude mice model and treatment strategy. (B) Individual and (C) average tumor growth curves in each treatment group. (D) Tumor inhibition rate and tumor weight toward MCF-7 tumor-bearing mice after treatment with Ctrl, siCDC20, HRNP/siCtrl, or HRNP/siCDC20. (F) Western blot and quantitative analyses of CDC20 expression in the Ctrl and HRNP/siCDC20 group. Immunofluorescence staining of (G) CDC20, (H) cyclin B (H), and (I) Mcl-1 expression in MCF-7 tumor tissues of xenograft mice after treatment with Ctrl, siCDC20, HRNP/siCtrl, or HRNP/siCDC20. Scale bar is 50 μm. (J) TUNEL of tumor tissue sections to detect tumor apoptosis in each treatment group. Scale bar is 50 μm. The nuclei and apoptotic cells are stained blue and green, respectively. The statistical data are presented as mean ± SD (n = 5 for in vivo study, n = 3 for ex vivo study; ***P < 0.001).
CONCLUSIONS
In summary, this study proposed a unique and precise breast cancer therapy by silencing the hypoxia-correlated protumorigenic gene using the hypoxia-responsive siRNA nanoparticle. We revealed that CDC20 expression was upregulated in the tumor tissue of breast cancer patients and was correlated with tumor hypoxia via bioinformatics analyses. Our in vitro studies also demonstrated a higher level of CDC20 expression in breast cancer cells under hypoxic conditions. As a proof of concept, a NI-modified polypeptide-based HRNP was developed for hypoxia-responsive siRNA delivery to target CDC20 specifically. The HRNP/siCDC20 with a PEGylated surface and small size showed prolonged blood circulation, high tumor accumulation, highly effective CDC20 silencing, and significant suppression of tumor growth. This combination of dual hypoxia-targeting presents a promising and advanced strategy for precision cancer therapy.
Supplementary Material
ACKNOWLEDGMENTS
The study was in part supported by the National Institutes of Health (NIH) (CA200900) and David Koch-PCF Award in Nanotherapeutics. Y.L. (201606170191) and R.S. (201708320347) were supported by the China Scholarship Council. X.X. would like express thanks for the financial support from the International Scientific and Technological Cooperation Program from Guangdong Science and Technology Department (2018A050506033).
Footnotes
The authors declare the following competing financial interest(s): O.C.F. has a financial interest in Selecta Biosciences, Tarveda Therapeutics, and Seer.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.0c00757.
Additional experimental details and figures including a diagram, syntheses, CDC20 levels, 1H NMR spectra, GPC, Dh, luciferase silence and cytotoxicity, FCM profiles, CLSM images, GSEA output, body weight changes, immunofluorescence staining, TUNEL staining, H&E staining, ALT and BUN levels, and tumor volume data (PDF)
Contributor Information
Yujing Li, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States; School of Life Science, Jilin University, Changchun 130012, P. R. China.
Jianxun Ding, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States.
Xiaoding Xu, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States; Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, P. R. China.
Run Shi, Department of Radiation Oncology, University Hospital, LMU Munich, Munich 81379, Germany.
Phei Er Saw, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States; Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou 510120, P. R. China.
Junqing Wang, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States.
Shirley Chung, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States.
Wenliang Li, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States.
Bader M. Aljaeid, King Abdulaziz University, Jeddah 21589, Saudi Arabia
Robert J. Lee, School of Life Science, Jilin University, Changchun 130012, P. R. China; Division of Pharmaceutics and Pharmacology, College of Pharmacy, The Ohio State University, Columbus, Ohio 43210, United States.
Wei Tao, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United Statess.
Lesheng Teng, School of Life Science, Jilin University, Changchun 130012, P. R. China.
Omid C. Farokhzad, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States.
Jinjun Shi, Center for Nanomedicine and Department of Anesthesiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States.
REFERENCES
- (1).Petrova V; Annicchiarico-Petruzzelli M; Melino G; Amelio I The Hypoxic Tumour Microenvironment. Oncogenesis 2018, 7 (1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Stewart GD; Ross JA; McLaren DB; Parker CC; Habib FK; Riddick AC The Relevance of a Hypoxic Tumour Microenvironment in Prostate Cancer. BJU Int. 2010, 105 (1), 8–13. [DOI] [PubMed] [Google Scholar]
- (3).Semenza GL Defining the Role of Hypoxia-Inducible Factor 1 in Cancer Biology and Therapeutics. Oncogene 2010, 29 (5), 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Rankin EB; Giaccia AJ The Role of Hypoxia-Inducible Factors in Tumorigenesis. Cell Death Differ. 2008, 15 (4), 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Harris AL Hypoxia—A Key Regulatory Factor in Tumour Growth. Nat. Rev. Cancer 2002, 2 (1), 38–47. [DOI] [PubMed] [Google Scholar]
- (6).Denko NC; Fontana LA; Hudson KM; Sutphin PD; Raychaudhuri S; Altman R; Giaccia AJ Investigating Hypoxic Tumor Physiology through Gene Expression Patterns. Oncogene 2003, 22 (37), 5907–5914. [DOI] [PubMed] [Google Scholar]
- (7).Sun M; Wang K; Oupicky D Advances in Stimulus-Responsive Polymeric Materials for Systemic Delivery of Nucleic Acids. Adv. Healthcare Mater 2018, 7, 1701070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Thambi T; Park JH; Lee DS Hypoxia-Responsive Nanocarriers for Cancer Imaging and Therapy: Recent Approaches and Future Perspectives. Chem. Commun 2016, 52 (55), 8492–8500. [DOI] [PubMed] [Google Scholar]
- (9).Wilson WR; Hay MP Targeting Hypoxia in Cancer Therapy. Nat. Rev. Cancer 2011, 11 (6), 393–410. [DOI] [PubMed] [Google Scholar]
- (10).Thambi T; Deepagan VG; Yoon HY; Han HS; Kim SH; Son S; Jo DG; Ahn CH; Suh YD; Kim K; Kwon IC; Lee DS; Park JH Hypoxia-Responsive Polymeric Nanoparticles for Tumor-Targeted Drug Delivery. Biomaterials 2014, 35 (5), 1735–1743. [DOI] [PubMed] [Google Scholar]
- (11).Yu J; Zhang Y; Ye Y; DiSanto R; Sun W; Ranson D; Ligler FS; Buse JB; Gu Z Microneedle-Array Patches Loaded with Hypoxia-Sensitive Vesicles Provide Fast Glucose-Responsive Insulin Delivery. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (27), 8260–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Qian C; Yu J; Chen Y; Hu Q; Xiao X; Sun W; Wang C; Feng P; Shen QD; Gu Z Light-Activated Hypoxia-Responsive Nanocarriers for Enhanced Anticancer Therapy. Adv. Mater 2016, 28 (17), 3313–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Son S; Rao NV; Ko H; Shin S; Jeon J; Han HS; Nguyen VQ; Thambi T; Suh YD; Park JH Carboxymethyl Dextran-Based Hypoxia-Responsive Nanoparticles for Doxorubicin Delivery. Int. J. Biol. Macromol 2018, 110, 399–405. [DOI] [PubMed] [Google Scholar]
- (14).Perche F; Biswas S; Wang T; Zhu L; Torchilin VP Hypoxia-Targeted siRNA Delivery. Angew. Chem., Int. Ed 2014, 53 (13), 3362–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kang L; Fan B; Sun P; Huang W; Jin M; Wang Q; Gao Z An Effective Tumor-Targeting Strategy Utilizing Hypoxia-Sensitive siRNA Delivery System for Improved Antitumor Outcome. Acta Biomater. 2016, 44, 341–354. [DOI] [PubMed] [Google Scholar]
- (16).Wang Z; Wan L; Zhong J; Inuzuka H; Liu P; Sarkar FH; Wei W CDC20: A Potential Novel Therapeutic Target for Cancer Treatment. Curr. Pharm. Des 2013, 19 (18), 3210–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wan L; Tan M; Yang J; Inuzuka H; Dai X; Wu T; Liu J; Shaik S; Chen G; Deng J; Malumbres M; Letai A; Kirschner MW; Sun Y; Wei W APCCDC20 Suppresses Apoptosis through Targeting Bim for Ubiquitination and Destruction. Dev. Cell 2014, 29 (4), 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kidokoro T; Tanikawa C; Furukawa Y; Katagiri T; Nakamura Y; Matsuda K CDC20, A Potential Cancer Therapeutic Target, is Negatively Regulated by p53. Oncogene 2008, 27 (11), 1562–1571. [DOI] [PubMed] [Google Scholar]
- (19).Manchado E; Guillamot M; de Carcer G; Eguren M; Trickey M; Garcia-Higuera I; Moreno S; Yamano H; Canamero M; Malumbres M Targeting Mitotic Exit Leads to Tumor Regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55 α,δ Phosphatase. Cancer Cell 2010, 18 (6), 641–54. [DOI] [PubMed] [Google Scholar]
- (20).Sackton KL; Dimova N; Zeng X; Tian W; Zhang M; Sackton TB; Meaders J; Pfaff KL; Sigoillot F; Yu H; Luo X; King RW Synergistic Blockade of Mitotic Exit by Two Chemical Inhibitors of the APC/C. Nature 2014, 514 (7524), 646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wang L; Zhang J; Wan L; Zhou X; Wang Z; Wei W Targeting CDC20 as a Novel Cancer Therapeutic Strategy. Pharmacol. Ther 2015, 151, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Mootha VK; Lindgren CM; Eriksson KF; Subramanian A; Sihag S; Lehar J; Puigserver P; Carlsson E; Ridderstrale M; Laurila E; Houstis N; Daly MJ; Patterson N; Mesirov JP; Golub TR; Tamayo P; Spiegelman B; Lander ES; Hirschhorn JN; Altshuler D; Groop LC PGC-1α-Responsive Genes Involved in Oxidative Phosphorylation are Coordinately Downregulated in Human Diabetes. Nat. Genet 2003, 34, 267–273. [DOI] [PubMed] [Google Scholar]
- (23).Subramanian A; Tamayo P; Mootha VK; Mukherjee S; Ebert BL; Gillette MA; Paulovich A; Pomeroy SL; Golub TR; Lander ES; Mesirov JP Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. U. S. A 2005, 102 (43), 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Karra H; Repo H; Ahonen I; Loyttyniemi E; Pitkanen R; Lintunen M; Kuopio T; Soderstrom M; Kronqvist P CDC20 and Securin Overexpression Predict Short-Term Breast Cancer Survival. Br. J. Cancer 2014, 110 (12), 2905–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Xu X; Xie K; Zhang XQ; Pridgen EM; Park GY; Cui DS; Shi J; Wu J; Kantoff PW; Lippard SJ; Langer R; Walker GC; Farokhzad OC Enhancing Tumor Cell Response to Chemotherapy through Nanoparticle-Mediated Codelivery of siRNA and Cisplatin Prodrug. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (46), 18638–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Xu X; Saw PE; Tao W; Li Y; Ji X; Yu M; Mahmoudi M; Rasmussen J; Ayyash D; Zhou Y; Farokhzad OC; Shi J Tumor Microenvironment-Responsive Multistaged Nanoplatform for Systemic RNAi and Cancer Therapy. Nano Lett. 2017, 17 (7), 4427–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Liu Y; Ji X; Tong WWL; Askhatova D; Yang T; Cheng H; Wang Y; Shi J Engineering Multifunctional RNAi Nanomedicine to Concurrently Target Cancer Hallmarks for Combinatorial Therapy. Angew. Chem., Int. Ed 2018, 57 (6), 1510–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.