Abstract
The present study aimed to investigate the expression of serum exosomal miR-23b-3p in non-small cell lung cancer (NSCLC) and to determine its diagnostic efficacy for NSCLC. From October, 2017 to October, 2019, 80 patients with NSCLC, 60 patients with pneumonia and 30 healthy subjects undergoing physical examination were enrolled at the People's Hospital of Yangzhong City. Serum samples were collected from the 3 groups of patients. The expression of miR-23b-3p in exosomes was detected by RT-qPCR. The Chi-squared test was used to analyze the expression level of miR-23b-3p in exosomes, and the patients with NSCLC were divided into 2 groups according to the expression level. The association between the patient clinicopathological parameters and receiver operating characteristic (ROC) curves was used to evaluate the diagnostic efficacy of serum exosomal miR-23b-3p in NSCLC. The expression level of serum exosomal miR-23b-3p in the patients with NSCLC was significantly higher than that in patients with pneumonia (t=10.332, P<0.001) and healthy subjects (t=12.810, P<0.001); serum exosomal miR-23b-3p was significantly associated with tumor size, depth of invasion, liver metastasis and TNM stage (P<0.05). The area under the curve (AUC) for miR-23b-3p was 0.915 (95% CI, 0.84-0.92), the optimal relative expression of miR-23b-3p was 3.46, the sensitivity of diagnosis was 87.4%, and the specificity was 93.8%, all higher than that of carcinoembryonic antigen (CEA). The ROCAUC of NSCLC was 0.645 (95% CI, 0.641-0.772) and for Cyfra21-1 it was 0.745 (95% CI, 0.701-0.812). Compared with the patients with pneumonia and the healthy subjects, the patients with NSCLC exhibited a higher level of serum exosomal miR-23b-3p. On the whole, these findings indicate that miR-23b-3p has a higher clinical diagnostic efficacy and may thus be a potential biomarker for the early diagnosis of NSCLC.
Keywords: serum exosomes, miR-23b-3p, non-small cell lung cancer, tumor markers
Introduction
Lung cancer is one of the most common malignant tumors, and its incidence and mortality rank first in China (1). Non-small cell lung cancer (NSCLC) is the most common pathological subtype, accounting for approximately 80% of the total number of lung cancer cases. The early symptoms of NSCLC are not obvious and the diagnosis is difficult, leading to the fact that the majority of patients are already in the middle and late stages of the disease at the time of diagnosis, thus severely threating the efficacy of surgical treatment (2,3). The 5-year survival rate of patients with stage I NSCLC is 80%, while that of patients with advanced NSCLC is only 15%, and the post-operative recurrence rate is very high (4). Therefore, it is of utmost clinical significance to identify specific tumor markers for the early diagnosis and treatment of NSCLC (5).
MicroRNAs (miRNAs or miRs) are endogenous non-coding small-molecule RNAs expressed in various types of tissues, and can be used for the regulation of the proliferation, differentiation and apoptosis of cells (6). Exosomes are a type of membrane vesicle (30–150 nm) containing a variety of RNA and proteins, which can be secreted under both normal and pathological conditions. Exosomes are one of the main vectors of miRNA transport and can stably exist in the circulatory system (7). There are a number of miRNAs with a dysregulated expression in patients with NSCLC. These miRNAs can be released into the circulation through exosome release. Serum exosomal miRNA can become a tumor biomarker of NSCLC (8). miR-23b-3p is one of the newly discovered and identified miRNAs, which is highly expressed in colon cancer, liver cancer and gastric cancer (9–11). In the present study, the expression of miR-23b-3p in serum exosomes in NSCLC, patients with pneumonia and healthy subjects was compared and analyzed, and the diagnostic efficacy of miR-23b-3p in serum exosomes in NSCLC was discussed.
Patients and methods
Patient information
The present study was approved by the Ethics Committee of the People's Hospital of Yangzhong City. Patients who participated in this research, signed the informed consent and had complete clinical data. A total of 80 patients with NSCLC and 60 patients with pneumonia were included in the present study. They were admitted to the People's Hospital of Yangzhong City from October, 2017 to October, 2019. During the same period, 30 healthy subjects in the physical examination center were included as the controls. The inclusion criteria were as follows: All patients with NSCLC were confirmed by histopathological examination, and staging was conducted according to the latest TNM stage 8 version of lung cancer (12). All patients with pneumonia were confirmed by clinical examination and did not receive antibiotics or other relevant treatment prior to specimen collection. The healthy subjects were those who had a regular medical examination in the physical examination center of our hospital for >2 years and did not have tumors in any part of the body. The exclusion criteria were as follows: Patients with NSCLC and pneumonia with incomplete clinical data, or healthy subjects with incomplete physical examination results. A total of 80 patients with NSCLC, 60 patients with pneumonia and 30 healthy subjects were included. Among the 80 patients with NSCLC, 54 were male and 26 were female. The mean age was (51.67±4.96) years. Smoking was found in 47 cases, accounting for 58.75%. There were 41 cases of adenocarcinoma and 39 cases of squamous cell carcinoma. As regards the clinical stage, 50 patients had stage I–II disease and 30 patients had stage III–IV disease. There were 49 cases of lymph node metastasis. Among the 60 patients with pneumonia, 36 were males and 24 were females. The mean age was (51.23±4.42) years. There were 31 smokers, accounting for 51.67%. Among the 30 healthy patients, 17 were males and 13 were females. The average age was (50.14±3.57) years. There were 17 smokers, accounting for 56.67%. No statistically significant differences were found in sex, age and smoking status between the patients with NSCLC and those with pneumonia and the healthy subjects (P>0.05).
Instruments and reagents
The Jem-100cx II transmission electron microscope was purchased from Hitachi, Ltd., and the nano-particle tracking analyzer Nanosight NS300 was from Malvern Instruments. The spectrophotometer was from Bole Life Medicine Products (Shanghai) Co., Ltd.; the exosome extraction kit was obtained from Applied Biosystems, Inc.. The 7500 real-time time PCR instrument was from Applied Biosystems; Thermo Fisher Scientific, Inc.. The reverse transcription kit and qPCR Kit from also from Applied Biosystems; Thermo Fisher Scientific, Inc., and the total RNA extraction kit and (TRIzol) reagent were from Invitrogen; Thermo Fisher Scientific, Inc.
Collection of blood samples
Fasting venous blood was collected from all patients and the healthy controls. Serum was obtained following centrifugation and stored at −80°C for analysis. At the same time, 2 ml blood samples were collected from each subject and sent to the nuclear medicine department for carcinoembryonic antigen (CEA) and Cyfra21-1 detection (the normal upper limit of CEA and Cyfra21-1 as indicated in the kit instructions was 5 and 7 µg/l, respectively).
Extraction and identification of serum exosomes
A total of 400 µl serum samples were gradually melted on an ice surface. The exosome extraction kit from Applied Biosystems, Inc. was used and the serum exosomes were extracted strictly according to the instructions provided with the kit. The exosomal samples were placed under an electron microscope to observe the morphology of the particles. The exosomal samples were diluted 5,000-, 10,000- or 20,000-fold to yield concentrations between 108 and 109/ml. The NanoSight NS300 instrument (Malvern Panalytical) was used for detection, the threshold was set to 5, and all detection parameters were kept unaltered during the detection process.
Detection of miR-23b-3p in exosomes
The same amount (100 µl) of exosomes was used for each sample. Total RNA was extracted from the exosomal samples using TRIzol reagent and purified by a silica gel adsorption column (Life Technologies; Thermo Fisher Scientificc, Inc.). cDNA was obtained by reverse transcription. The reaction conditions were as follows: 16°C for 15 min, 42°C for 60 min, 85°C for 5 min, and 4°C for 5 min. miR-23b-3p expression in exosomes was detected quantitatively by fluorescent dye participation method [SYBR-Green, Tiangen Biotech (Beijing) Co., Ltd.]. The primer sequences were as follows: miR-23b-3p forward, 5′-TCACGGTCCAGTTTTCCCAG-3′ and reverse, 5′-GGAAGGAGGCAAATCCAGCT-3′; miRNA-39 forward, 5′-TGGAAAGGACGAAACACCGT-3′ and reverse, 5′-ATTTGCGTGTCATCCTTGCG-3. The expression level of miR-23b-3p in exosomes was calculated using the 2−ΔΔCq method (13) and miRNA-39 was used as the external reference gene.
Statistical analysis
SPSS 18.0 statistical software was used to analyze the data. The measurement data are expressed as the means ± standard deviation (SD). One-way variance analysis followed by the LSD test was used for comparisons between multiple groups. The counting data were expressed by case number and rate (%), and the Chi-squared (χ2) test was used for comparisons between groups. The diagnostic efficacy of serum exosomal miR-23b-3p, serum CEA and Cyfra21-1 in NSCLC was evaluated by receiver operating characteristic (ROC) curve analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of serum exosomes
The structure and morphology of the serum exosomes extracted from the samples of the 3 groups of study subjects were relatively similar (data not shown). Under an electron microscope, the serum exosome sample from a patient with NSCLC exhibited a lipid bilayer membrane with cystic round particles with a diameter of approximately 100 nm (Fig. 1). NanoSight revealed that the serum exosome particles were uniformly dispersed, and the diameter of the exosome body was approximately 50–180 nm, the distribution curve exhibited a single peak normal distribution and the peak value was approximately 104 nm (Fig. 2).
Figure 1.
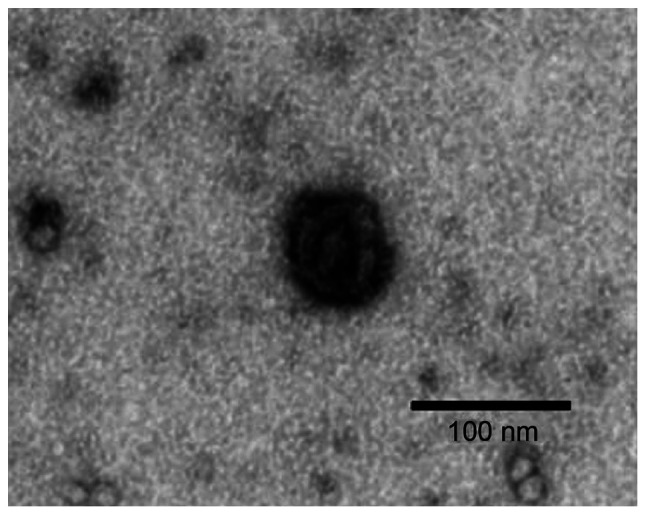
Morphology of serum exosomal particles (from a patient with non-small cell lung cancer) under an electron microscope (magnification, ×50,000). Under an electron microscope, exosomes exhibited a round double-layer membrane and coated particles, approximately 100 nm in diameter.
Figure 2.
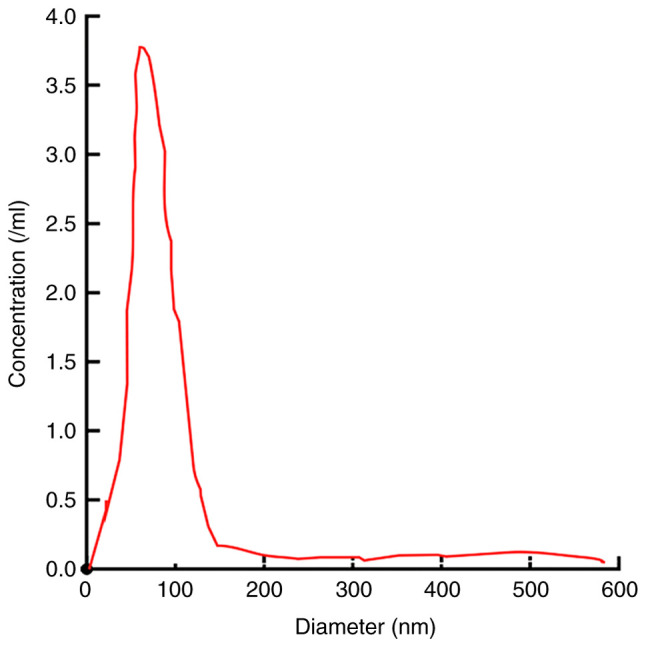
Detection of diameter and concentration of exosomes by NanoSight. The diameter of exosomes particles was 70–180 nm. The distribution was even, and there was a single peak, 104 nm.
Expression of miR-23b-3p in serum exosomes
A significant difference was observed in the expression of miR-23b-3p in serum exosomes from the patients with NSCLC, the patients with pneumonia and the healthy subjects (F=16.202, P<0.01). The expression level of serum exosomal miR-23b-3p in patients with NSCLC was significantly higher than that in patients with pneumonia (t=10.332, P<0.001) and healthy subjects (t=12.810, P<0.001; Fig. 3).
Figure 3.
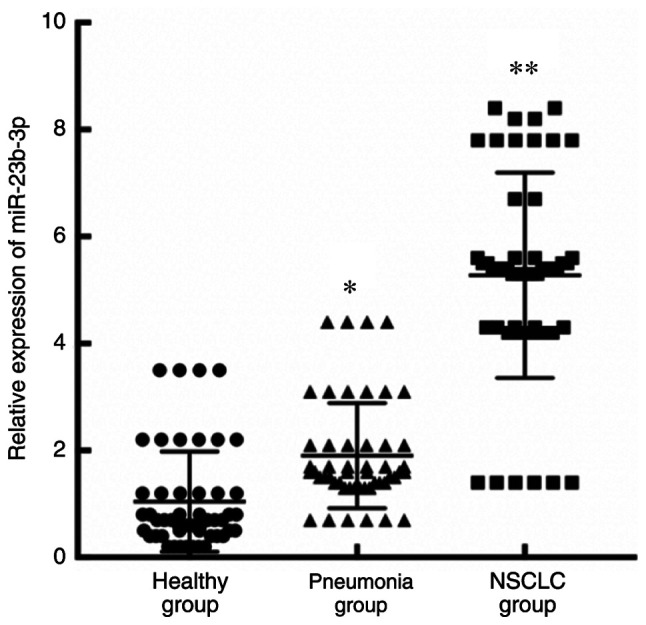
Relative expression of miR-23b-3p in serum exosomes from the patients in the 3 groups. The same amount (100 µl) of exosomes was used for each sample. NSCLC, non-small cell lung cancer. *P<0.05 and **P<0.05 vs. healthy group and/or pneumonia group.
Association between serum exosomal miR-23b-3p and clinicopathological parameters of patients with lung cancer
The 80 patients with NSCLC were divided into the high expression group and low expression group according to the relative expression level of miR-23b in exosomes (median relative expression level of mir-23b-3p in exosomes was 3.98). There were 40 cases with a high exosomal miR-23b-3p expression and 40 cases with a low exosomal miR-23b-3p expression. There was a significant association between serum exosomal miR-23b-3p and tumor size, depth of invasion, liver metastasis and TNM stage (Table I).
Table I.
Association between serum exosomal miR-23b-3p expression and clinicopathological parameters of patients with NSCLC.
| Variable | miR-23b-3p low expression group | miR-23b-3p high expression group | P-value |
|---|---|---|---|
| Sex | 0.400 | ||
| Male | 28 | 26 | |
| Female | 12 | 14 | |
| Size of tumors (cm) | 0.019 | ||
| <5 | 22 | 20 | |
| ≥5 | 18 | 20 | |
| Differentiation degree | 0.161 | ||
| High/moderate | 25 | 26 | |
| Poor | 15 | 14 | |
| Depth of invasion | 0.029 | ||
| pT1 | 10 | 11 | |
| ≥pT2 | 30 | 29 | |
| Lymphatic metastasis | 0.136 | ||
| No | 15 | 16 | |
| Yes | 25 | 24 | |
| Liver metastasis | 0.032 | ||
| No | 32 | 37 | |
| Yes | 8 | 3 | |
| Peritoneal carcinomatosis | 0.248 | ||
| No | 31 | 30 | |
| Yes | 9 | 10 | |
| TNM staging | 0.034 | ||
| I | 13 | 12 | |
| II | 12 | 13 | |
| III | 8 | 7 | |
| IV | 7 | 8 |
Diagnostic efficacy of miR-23b-3p in serum exosomes in NSCLC
ROC curve analysis was used to evaluate the diagnostic efficacy of serum exosomal miR-23b-3p in NSCLC. The area under the curve of serum exosomal miR-23b-3p was 0.915 (95% CI: 0.84-0.92). The optimal critical value of miR-23b-3p relative expression was 3.46, the diagnostic sensitivity was 87.4%, and the specificity was 93.8%. The area under the ROC curve (AUC) for serum CEA, a traditional tumor marker, was 0.645 (95% CI, 0.641-0.772) in the diagnosis of NSCLC. The critical value of CEA relative expression was 7.46, the diagnostic sensitivity was 67.4% and the specificity was 66.8%. The AUC of serum Cyfra21-1 for the diagnosis of NSCLC was 0.745 (95% CI, 0.701-0.812), the critical value of Cyfra21-1 relative expression was 8.56, and the diagnostic sensitivity and specificity were 70.4 and 71.8%, respectively (Fig. 4).
Figure 4.
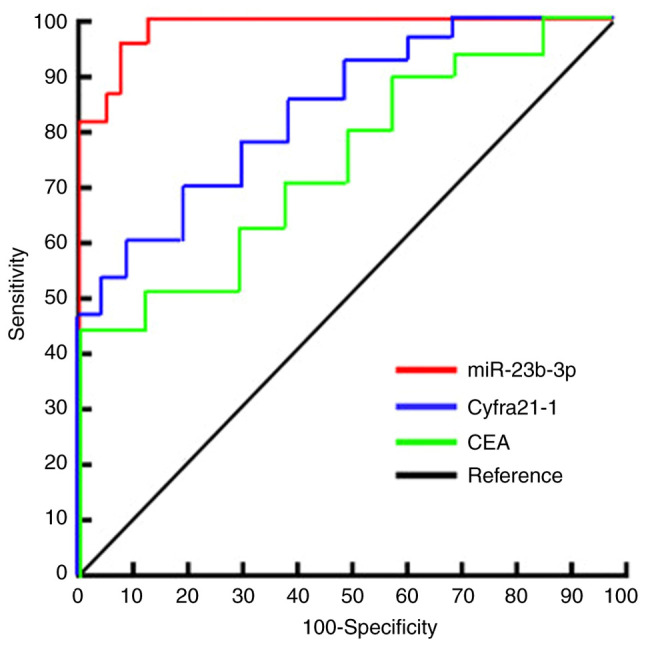
miR-23b-3p and CEA in serum exosomes and ROC curve analysis for the diagnosis of NSCLC. NSCLC, non-small cell lung cancer; CEA, carcinoembryonic antigen.
Discussion
NSCLC is the most common malignant tumor in China, which severely threatens the health of residents and places a heavy burden on families and society. The search for non-invasive serum markers for patients with NSCLC is of utmost importance for the early diagnosis and follow-up evaluation of patients with lung cancer (14). Surgery combined with simultaneous radiotherapy and chemotherapy can improve the clinical cure rate of early-stage NSCLC; however, there is still a lack of effective treatment for patients with advanced disease, whose survival rate is <15%. No significant improvements have been made in this field for a number of years (15). Thus, it is of utmost clinical significance to explore early NSCLC diagnostic markers.
miRNAs, short chain non-coding RNAs, play a key role in the occurrence and progression of cancer, and have gradually become molecular markers for cancer diagnosis and prognosis (16,17). In recent years, a large number of studies have indicated that the expression of miRNAs in tumor tissues or cells differs from that in normal tissues, and this differential expression is closely related to tumorigenesis and progression. miRNAs are potential molecular markers of tumors and can play an important role in the early diagnosis and prognosis of tumors (18,19). It has been found that a variety of miRNAs, such as miR-17, miR-190b, miR-19a, miR-19b, miR-26b and miR-375, are related to the occurrence and progression of NSCLC, and have certain diagnostic value for NSCLC (20,21). A recent study (22) also demonstrated that serum miRNA is expected to be a diagnostic marker of NSCLC.
Although serum miRNAs are valuable for the diagnosis of clinical NSCLC, there are certain shortcomings: The composition of serum is complex and the source of miRNAs is unknown, which affects the specificity of the detection results. miRNAs in serum are also affected by a number of enzymes, resulting in poor stability (23). Recently, it has been found that there is a certain difference in the expression profiles between serum miRNAs and serum exosomes. Compared with serum miRNAs, serum exosomes are less disturbed and have better stability (24). miRNAs exist stably in serum exosome bodies and are less affected by various enzymes in serum. The samples can be obtained in a non-invasive manner, which renders serum exosomal miRNAs a non-invasive and reliable tumor biomarkers (25).
In the present study, exosomes in serum were directly extracted using a serum exosome extraction kit. Examination by a transmission electron microscope and a NasoSight instrument revealed that there was a large number of high-abundance exosome particles in the samples of the 3 groups. At present, there are 4 main methods reported in the literature for the extraction and purification of exosomes: Ultracentrifugation, immunomagnetic bead method, polyethylene glycol precipitation method and the kit method (26). In the present study, the kit from Applied Biosystems, Inc. was used to extract and confirm the high purity and abundance of serum exosomes, which provided the basis for subsequent research (26). In this study, it was found that the purified serum samples contained a large number of exosome particles. The expression of miR-23b-3p in serum exosomes of patients with NSCLC was significantly higher than that of patients with pneumonia and healthy controls. ROC curve analysis was used to evaluate the diagnostic efficacy of miR-23b-3p in NSCLC. The results suggested that serum exosomal miR-23b-3p was more effective in the diagnosis of NSCLC. CEA and Cyfra21-1 are common traditional biomarkers in the diagnosis of NSCLC and for monitoring tumor recurrence (27). In the present study, it was found that the AUC of miR-23b-3p was greater than that of CEA and Cyfra21-1, and the sensitivity and specificity of miR-23b-3p were greater than those of CEA and Cyfra21-1. In comparison, miR-23b-3p in serum exosomes was superior to the traditional tumor markers, CEA and Cyfra21-1, in the diagnosis of NSCLC.
The results of the present study demonstrated that there was a statistically significant association between serum exosomal miR-23b-3p and tumor size, depth of invasion, liver metastasis and TNM stage in patients with NSCLC, suggesting that exosomal miR-23b-3p was closely related to the clinical progress of NSCLC. It has been reported (8–10) that miR-23b-3p plays an important role in the progression of colon cancer, liver cancer and gastric cancer, which is consistent with the results of the present study. It has been found that miR-23b-3p is an important miRNA that plays the role of a proto-oncogene. The results of whole gene sequencing of cancer and normal tissues in patients with NSCLC have demonstrated that the expression in NSCLC tissues seems to be significantly higher than that in healthy cancer tissues (28). A high expression of miR-23b-3p was considered to be a molecular marker of a poor prognosis in a single sample of 114 NSCLC patients (28). In the present study, the expression of miR-23b-3p in the serum of patients with NSCLC was not investigated. A recent study reported that a high expression of miR-23b-3p in serum exosomes was a marker of a poor prognosis of patients with pancreatic cancer (29). Although its involvement in the development and progression of pancreatic cancer has not yet been elucidated, it may promote the progress of pancreatic cancer by regulating annexin A2 expression (30). The above-mentioned results indicate that miR-23b-3p may regulate the expression of different related target genes in different tumor types, and may generally play a role similar to that of a proto-oncogene. The results of the present study initially confirmed that serum exosomal miRNA was closely related to the occurrence and progression of NSCLC. Since the detection of serum exosomal miRNA can be obtained in a non-invasive manner, it may become a non-invasive diagnostic marker of NSCLC.
In conclusion, the present study demonstrates that miR-23b-3p is highly expressed in serum exosomes of patients with NSCLC and has a high diagnostic efficacy for NSCLC; thus, it may become a novel diagnostic marker for NSCLC. However, further studies are required with an increased number of samples in order to further explore the asociation between serum exosomal miRNA and NSCLC, so as to provide reference for the early clinical diagnosis and treatment of NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JW and HX conceived and designed the study. ZZ and JG were responsible for the collection and analysis of the experimental data. MZ interpreted the data and drafted the manuscript. ZM performed the detection of miR-23b-3p in exosomes and revised the manuscript critically for important intellectual content. JW wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the People's Hospital of Yangzhong City. Patients who participated in this research, signed the informed consent and had complete clinical data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhang ML, Peng P, Wu CX, Gong YM, Zhang SW, Chen WQ, Bao PP. Report of breast cancer incidence and mortality in China registry regions, 2008–2012. Zhonghua Zhong Liu Za Zhi. 2019;41:315–320. doi: 10.3760/cma.j.issn.0253-3766.2019.04.013. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 2.He Y, Li D, Shan B, Liang D, Shi J, Chen W, He J. Incidence and mortality of esophagus cancer in China, 2008–2012. Chin J Cancer Res. 2019;31:426–434. doi: 10.21147/j.issn.1000-9604.2019.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caballero Vázquez A, García Flores P, Romero Ortiz A, Del Moral RG, Alcázar-Navarrete B. Changes in non-small cell lung cancer diagnosis, molecular testing and prognosis 2011–2016. J Thorac Dis. 2018;10:5468–5475. doi: 10.21037/jtd.2018.08.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez FG, Kosinski AS, Furnary AP, Onaitis M, Kim S, Habib RH, Tong BC, Cowper P, Boffa D, Jacobs JP, et al. Differential effects of operative complications on survival after surgery for primary lung cancer. J Thorac Cardiovasc Surg. 2018;155:1254–1264.e1. doi: 10.1016/j.jtcvs.2017.09.149. [DOI] [PubMed] [Google Scholar]
- 5.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 6.Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12–23. doi: 10.1016/j.critrevonc.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Chen R, Xu X, Qian Z, Zhang C, Niu Y, Wang Z, Sun J, Zhang X, Yu Y. The biological functions and clinical applications of exosomes in lung cancer. Cell Mol Life Sci. 2019;76:4613–4633. doi: 10.1007/s00018-019-03233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nigita G, Distefano R, Veneziano D, Romano G, Rahman M, Wang K, Pass H, Croce CM, Acunzo M, Nana-Sinkam P. Tissue and exosomal miRNA editing in non-small cell lung cancer. Sci Rep. 2018;8:10222. doi: 10.1038/s41598-018-28528-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grisard E, Coan M, Cesaratto L, Rigo I, Zandonà L, Paulitti A, Andreuzzi E, Rampioni Vinciguerra GL, Poletto E, Del Ben F, et al. Sleeping beauty genetic screen identifies miR-23b::BTBD7 gene interaction as crucial for colorectal cancer metastasis. EBioMedicine. 2019;46:79–93. doi: 10.1016/j.ebiom.2019.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang T, He X, Chen A, Tan K, Du X. LncRNA HOTAIR contributes to the malignancy of hepatocellular carcinoma by enhancing epithelial-mesenchymal transition via sponging miR-23b-3p from ZEB1. Gene. 2018;670:114–122. doi: 10.1016/j.gene.2018.05.061. [DOI] [PubMed] [Google Scholar]
- 11.Xian X, Tang L, Wu C, Huang L. miR-23b-3p and miR-130a-5p affect cell growth, migration and invasion by targeting CB1R via the Wnt/β-catenin signaling pathway in gastric carcinoma. Onco Targets Ther. 2018;11:7503–7512. doi: 10.2147/OTT.S181706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hattori A, Takamochi K, Oh S, Suzuki K. New revisions and current issues in the eighth edition of the TNM classification for non-small cell lung cancer. Jpn J Clin Oncol. 2019;49:3–11. doi: 10.1093/jjco/hyy142. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer. 2019;10:3–7. doi: 10.1111/1759-7714.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W, Sun K, Zheng R, Zeng H, Zhang S, Xia C, Yang Z, Li H, Zou X, He J. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Y, Zhang Y, Du L, Jiang X, Yan S, Duan W, Li J, Zhan Y, Wang L, Zhang S, et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol Oncol. 2018;12:648–658. doi: 10.1002/1878-0261.12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Yin Z, Fan J, Zhang S, Yang W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct Target Ther. 2019;4:47. doi: 10.1038/s41392-019-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallach S, Jantus-Lewintre E, Calabuig-Fariñas S, Montaner D, Alonso S, Sirera R, Blasco A, Usó M, Guijarro R, Martorell M, Camps C. MicroRNA profiling associated with non-small cell lung cancer: Next generation sequencing detection, experimental validation, and prognostic value. Oncotarget. 2017;8:56143–56157. doi: 10.18632/oncotarget.18603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu S, Kong H, Hou Y, Ge D, Huang W, Ou J, Yang D, Zhang L, Wu G, Song Y, et al. Two plasma microRNA panels for diagnosis and subtype discrimination of lung cancer. Lung Cancer. 2018;123:44–51. doi: 10.1016/j.lungcan.2018.06.027. [DOI] [PubMed] [Google Scholar]
- 21.Zaporozhchenko IA, Morozkin ES, Ponomaryova AA, Rykova EY, Cherdyntseva NV, Zheravin AA, Pashkovskaya OA, Pokushalov EA, Vlassov VV, Laktionov PP. Profiling of 179 miRNA expression in blood plasma of lung cancer patients and cancer-free individuals. Sci Rep. 2018;8:6348. doi: 10.1038/s41598-018-24769-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P, Yang D, Zhang H, Wei X, Ma T, Cheng Z, Hong Q, Hu J, Zhuo H, Song Y, et al. Early detection of lung cancer in serum by a panel of microRNA biomarkers. Clin Lung Cancer. 2015;16:313–319.e1. doi: 10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Xu Y, Liao X, Liao R, Zhang L, Niu K, Li T, Li D, Chen Z, Duan Y, Sun J. Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol. 2016;37:11927–11936. doi: 10.1007/s13277-016-5052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munagala R, Aqil F, Gupta RC. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumour Biol. 2016;37:10703–10714. doi: 10.1007/s13277-016-4939-8. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang YT, Huang YY, Zheng L, Qin SH, Xu XP, An TX, Xu Y, Wu YS, Hu XM, Ping BH, Wang Q. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int J Mol Med. 2017;40:834–844. doi: 10.3892/ijmm.2017.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang ZF, Wang M, Xu JL. Thymidine kinase 1 combined with CEA, Cyfra21-1 and NSE improved its diagnostic value for lung cancer. Life Sci. 2018;194:1–6. doi: 10.1016/j.lfs.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 28.Begum S, Hayashi M, Ogawa T, Jabboure FJ, Brait M, Izumchenko E, Tabak S, Ahrendt SA, Westra WH, Koch W, et al. An integrated genome-wide approach to discover deregulated microRNAs in non-small cell lung cancer: Clinical significance of miR-23b-3p deregulation. Sci Rep. 2015;5:13236. doi: 10.1038/srep13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen D, Wu X, Xia M, Wu F, Ding J, Jiao Y, Zhan Q, An F. Upregulated exosomic miR-23b-3p plays regulatory roles in the progression of pancreatic cancer. Oncol Rep. 2017;38:2182–2188. doi: 10.3892/or.2017.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei DM, Dang YW, Feng ZB, Liang L, Zhang L, Tang RX, Chen ZM, Yu Q, Wei YC, Luo DZ, Chen G. Biological effect and mechanism of the miR-23b-3p/ANXA2 axis in pancreatic ductal adenocarcinoma. Cell Physiol Biochem. 2018;50:823–840. doi: 10.1159/000494468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


