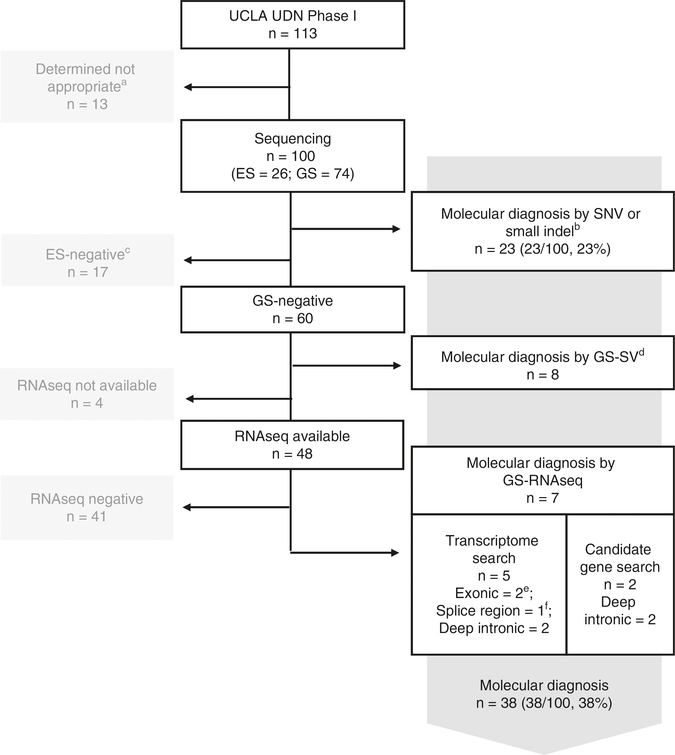
Fig. 1. Molecular diagnostic rate in the 113 Undiagnosed Diseases Network-University of California–Los Angeles (UDN-UCLA) clinical site cohort enrolled between July 2014 and August 2018.
aDetermined not appropriate for UDN genetic study after clinical evaluation. bSingle-nucleotide variant (SNV)/small indel variants within coding exons (includes essential splice site (+/−2 bp) variants) that are predicted to be nonsynonymous or loss-of-function: of the 23 probands, 9 were diagnosed with exome sequencing (35%; includes 1 proband who was diagnosed with a recurrent deep intronic pathogenic variant in COL6A1) and 14 with genome sequencing (19%). cExome-negative cases were removed from further analysis for this study due to the lack of DNA sequencing data in the noncoding genomic region. dSV: Structural variants affecting coding exons (includes mixed triploidy and repeat expansion). eVariants that are synonymous or in untranslated region (UTR). fVariants within +/−3 to +/−10 bp from the exon-intron boundaries. ES exome sequencing, GS genome sequencing.