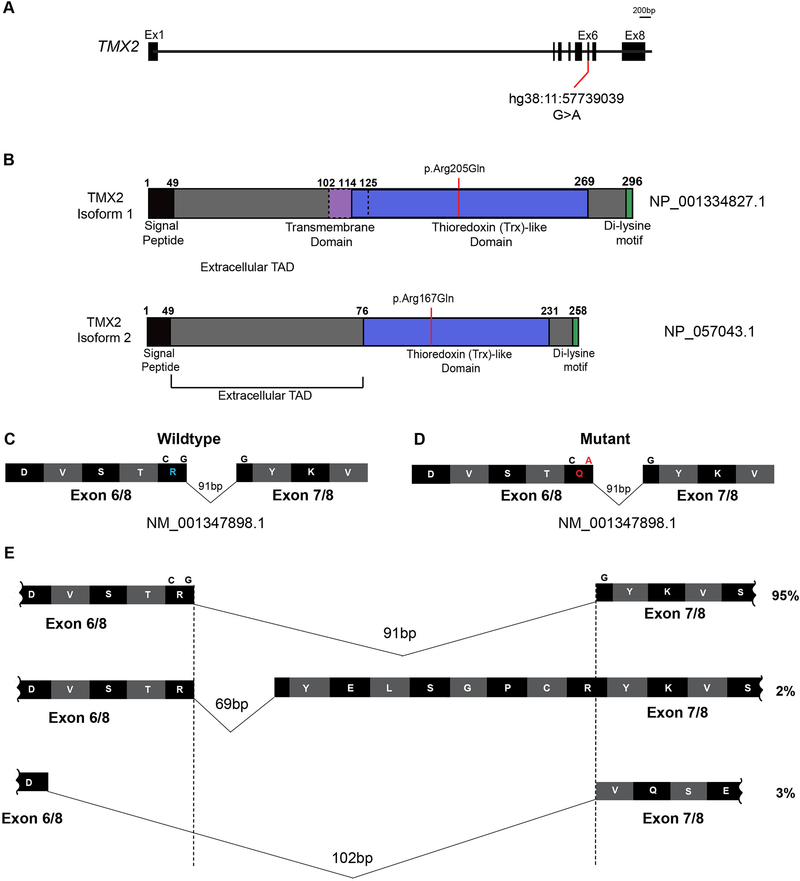
Figure 4. TMX2 gene and predicted protein isoforms.
(A) Schematic of TMX2 depicting the genomic map spanning eight exons. Red line indicates the position of the identified variant in exon six in all four families and coordinates within the cDNA map (RefSeq NM_001347898.1). (B) Schematic of the two protein-coding isoforms of Thioredoxin-related transmembrane protein 2 (TMX2) depicting the location of the ‘signal peptide’ (black), extracellular topological associated domain (TAD), transmembrane domain (purple), Thioredoxin (Trx)-like domain (blue), and Di-lysine motif (green). Isoform 2 is 38 amino acids shorter than isoform 1 due to absence of the transmembrane domain and part of the extracellular TAD. Red line indicates the position and coordinates of the amino acid change in affected individuals from all four families within the Trx-like domain. (C-D) Illustration of partial wildtype (C) and mutant (D) residues from exons six and seven surrounding the variant shown in blue and red in wildtype and mutant, respectively. (E) Schematic of the additional splice isoforms of the exon6–7 junction of TMX2, based on the UCSC Genome Browser with predicted percent existence of each isoform in human tissues from GenBank. The percentage of each isoform was calculated based on the average number of Expressed Sequence Tags (ESTs) available, displaying that particular isoform (e.g. BI764082, BG170274, and BI561678, respectively).