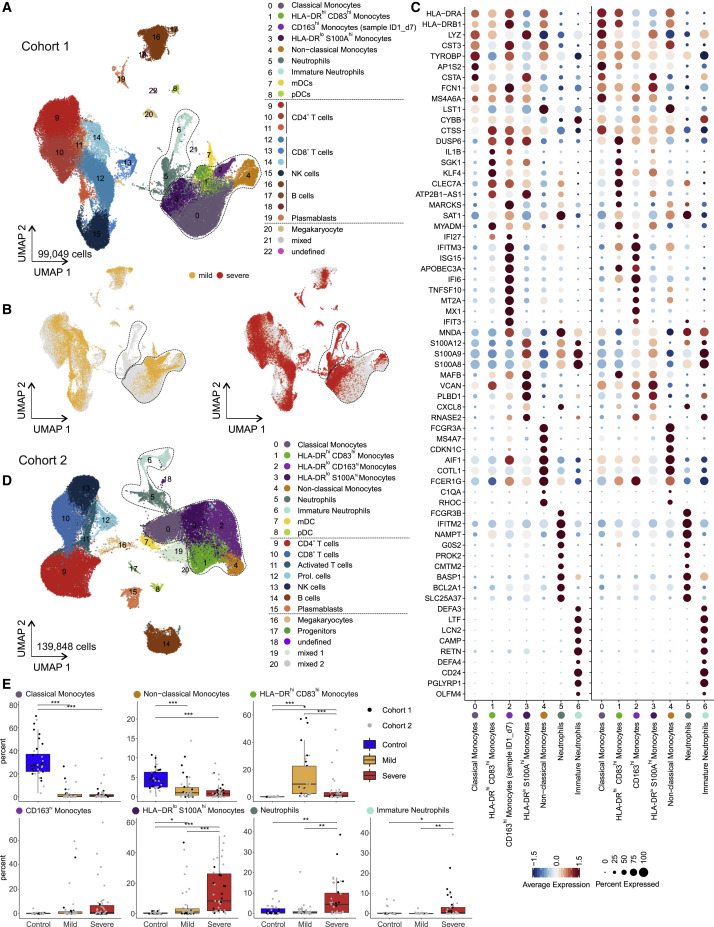
Figure 2.
scRNA-Seq of PBMC from Patients of the Two Independent Cohorts
(A) UMAP visualization of scRNA-seq profiles (10x, cohort 1) of 99,049 PBMC from 49 samples (8 mild, 10 severe patients, different time points) and 22 control samples colored according to cell type classification (Louvain clustering), reference-based cell-type annotation, and marker gene expression (Table S4).
(B) UMAP shown in (A) colored according to disease severity (yellow, mild COVID-19; red, severe COVID-19).
(C) Dot plots of the intersection of the top 20 marker genes sorted by average log fold change determined for the indicated myeloid cell subsets in the PBMC datasets of both cohorts.
(D) UMAP visualization of scRNA-seq profiles (BD Rhapsody, cohort 2) of 139,848 PBMCs (50 samples of 8 mild, 9 severe COVID-19; 14 samples of 13 controls; different time points), coloring as in (A) (see also Figure S2A and Table S4).
(E) Box and whisker plots (25–75 percentile) of percentages of cell subsets of total PBMC (per patient). Boxes are colored according to disease group and dots according to the respective cohort of the sample. Dirichlet-multinomial regression adjusted with the Benjamini-Hochberg method, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
See also Table S1.