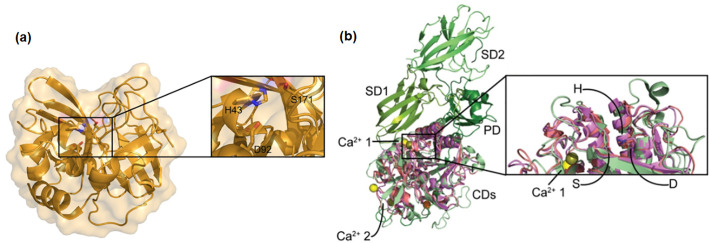
Fig. 6.
Crystal structures of keratinolytic enzymes of family S1 and S8. (a) Catalytic domain structure of ANISEP, a family S1 keratinase (accession number: BAM67011, PDB ID: 3WY8). The active site catalytic triad amino acids His43-Asp92- Ser171, situated in the upper part of the shallow cleft in the center, are magnified and displayed as sticks. (b) Structures of Fervidolysin (green), Proteinase K (violet), and MtaKer (salmon) family S8 keratinases with the active sites magnified and catalytic triad amino acids, His (H); Asp (D); and Ser (S) displayed as sticks. The CD domain corresponds to the catalytic domain; the PD domain is the propeptide domain; and SD1 and SD2 are β-sandwich domains. The CD domain of Fervidolysin is aligned with the structure of Proteinase K (accession number: 1205229A, PDB ID: 5WSL) and MtaKer (accession number: ARH33809, PDB ID: 5WSL). Proteinase K: Ca2+ ion = yellow, Fervidolysin: Ca2+ ion = orange, MtaKer Ca2+ ion = olive. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)