Figure 2.
The importance of reference database choice, design, and versioning in taxonomic profiling of clinical metagenomics samples
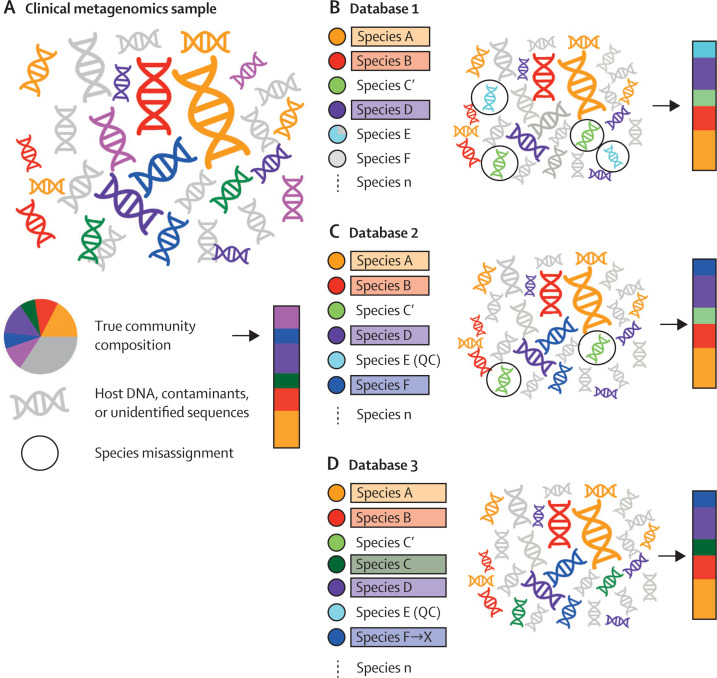
(A) Schematic representation of a typical clinical metagenomics sample with species assigned as coloured DNA and grey denoting DNA deriving from the host, contaminants, unidentified taxa, or taxa sequenced at low depth. The pie chart provides the full metagenomic composition with the bar providing the species composition excluding host DNA and contaminants. (B) Taxonomic profiling based on database 1. Species confidently assigned are highlighted by colours with unassigned species shown in grey. Using database 1, species A, B, and D are correctly assigned. Species that are misassigned are outlined with a circle. In this instance, sequences from species C are assigned to the closely related species C' because of the lack of a representative of species C in the reference database. Additionally, the reference database contains a partially contaminated sequence from species E, which is misassigned to contaminant sequences in the test clinical metagenomics sample. This affects the inference of species composition shown in the bar. (C) The addition of species F to database 2 allows assignment of a greater proportion of the species present in the original clinical metagenomics sample. Quality control and improvement of reference species E, now species E (QC), removes the spurious assignment of contaminant species. Species C is still misassigned to species C', its closest representative in the database. (D) Updating the reference database to include species C results in the correct assignment of sequences to species C rather than species C'. Species F is taxonomically reassigned to species X, leading to a change in the assigned species name despite no change in the data in the reference or query datasets. In all cases the pink sequences present in the original clinical metagenomics sample are not assigned as this species is not present in any of the three reference databases.