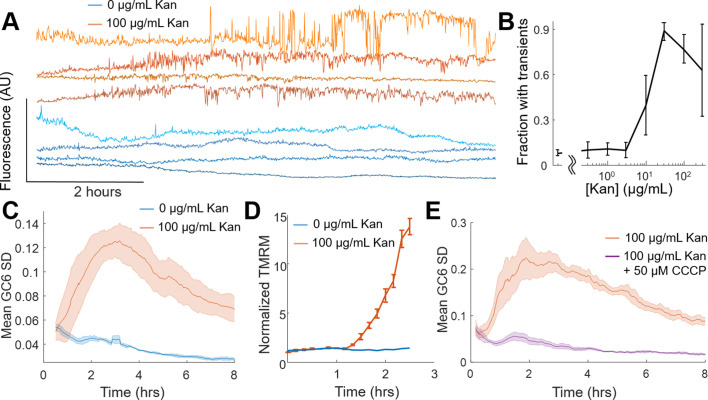
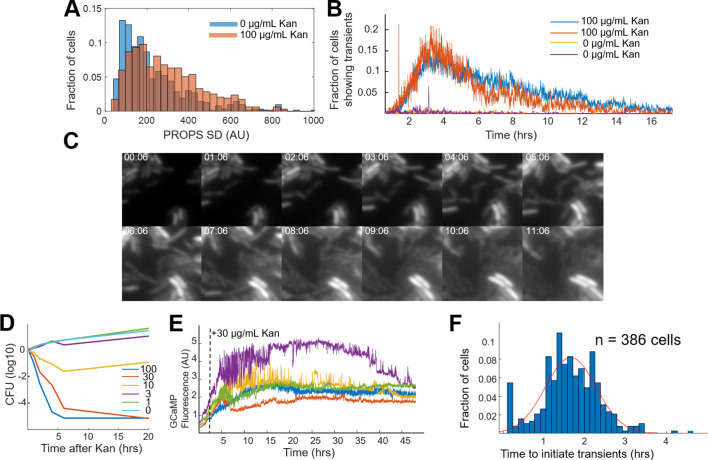
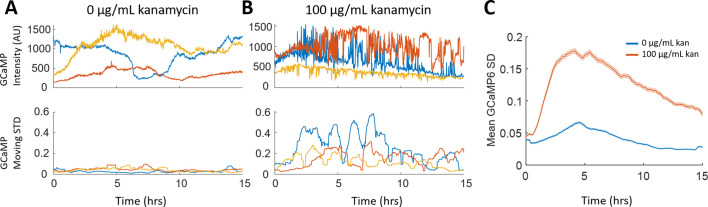
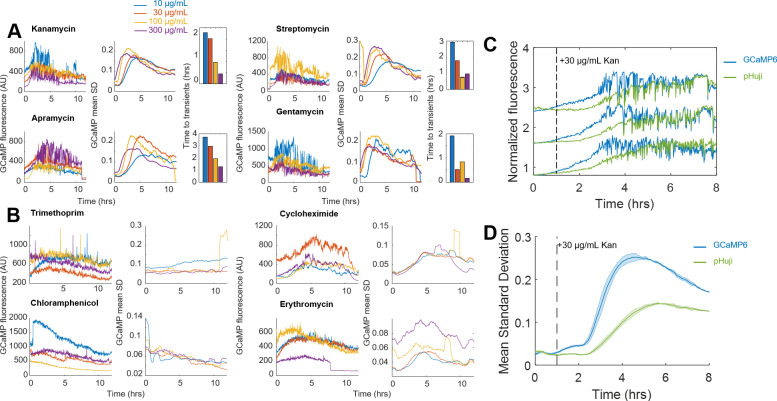
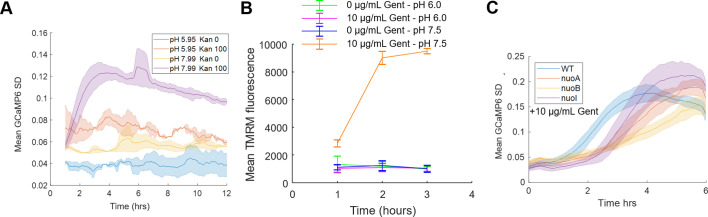
Figure 2. Voltage and calcium exhibit altered electrophysiological flux in response to aminoglycosides.
(A) Time traces of GCaMP6 fluorescence from single cells treated with 0 µg/mL (blue shades) and 100 µg/mL (orange shades) kanamycin. Individual cells display non-oscillatory transients. (B) The fraction of cells in a population of GCaMP6F expressing cells E. coli experiencing the transients in A at different concentrations of Kanamycin. The mean (line) and standard deviation (error bars) are shown for three biological replicates. (C) The average (solid line) and standard deviation (shading) of the moving GCaMP6f standard deviation (SD) over time from 0 µg/mL (blue) and 100 µg/mL kanamycin (orange) treated cells. (D) TMRM fluorescence from untreated (blue) or kanamycin treated (orange) (100 µg/mL, 2 hr) E. coli measured by cytometry. The average (line) and standard deviation (error bars) of three biological replicates are plotted. (E) The average (solid line) and standard deviation (shading) of the moving GCaMP6f Standard Deviation over time from 100 µg/mL kanamycin-treated cells in the absence (orange) or presence (purple) of 50 µM CCCP.