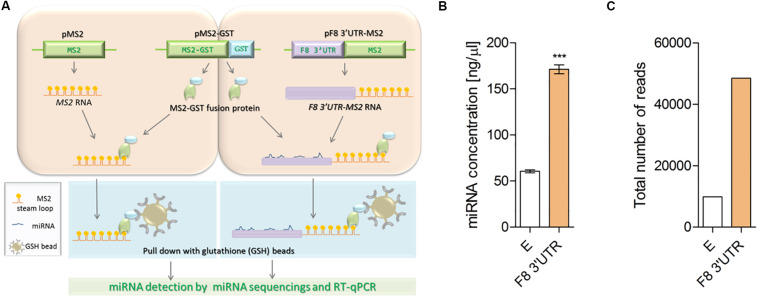
FIGURE 1.
Illustration of MS2 affinity purification method. (A) Plasmids: pMS2 (expressing control MS2 RNA, consisting of tandem 24 MS2 hairpins, orange), pMS2-GST (expressing a fusion protein that contains MS2 RNA-recognizing portion, green), and a region (GST, glutathione S-transferase, blue) that recognizes the affinity purification reagent glutathione-SH (GSH) and pMS2-F8 3′UTR [expressing the test RNA of interest (purple) tagged with 24 MS2 hairpins (F8 3′UTR-MS2)], were transfected into HEK-293T cells for expression of the RNAs and encoded protein. HEK-293T cells were transfected with (1) the plasmids to express control RNA (MS2 RNA) and reporter protein (MS2-GST) or (2) plasmids to express the experimental RNA of interest (F8 3′UTR-MS2 RNA) and the reporter protein (MS2-GST). After formation of the RNA-proteins (RNP) complexes, cells were lysed and the RNPs were affinity-purified by using GSH beads. Subsequently, RNA was isolated from the complexes and subjected to next generation sequencing to identify the bound microRNAs. (B) Affinity purification (average of three independent experiments) demonstrated higher concentration of miRNAs in MS2-tagged F8 3′UTR (E = 60.50 ± 3.10, N = 4; F8 3′UTR = 171.30 ± 9.66, N = 4) and (C) Total read counts detected in empty control (E = 9886) and MS2-tagged F8 3′UTR (F8 3′UTR = 48486) of pull-down samples. ***P < 0.001.