Figure 1.
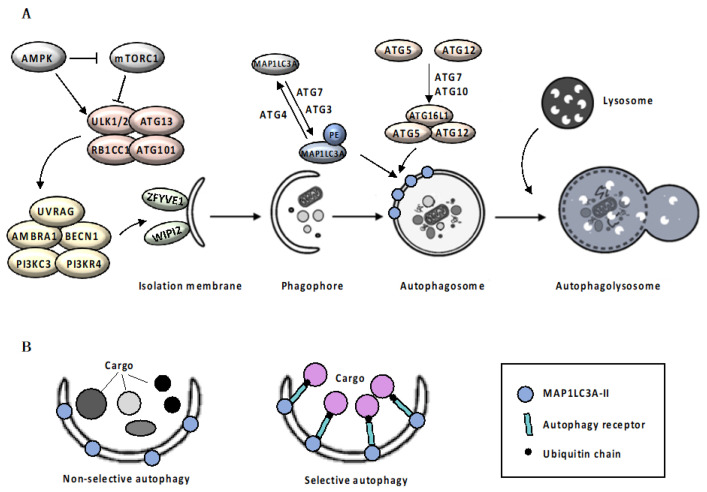
Autophagic Machinery. Box 1: Mechanism of autophagy. Autophagy initiates with the elongation of a precursor structure, called phagophore, surrounding cargo, to generate a double-membrane vesicle, called autophagosome [8,9]. Autophagosomes biogenesis is orchestrated by a subset of autophagy-related (ATG) proteins [10]. Upon autophagic stimuli, activation of AMPK (adenosine monophosphate (AMP)-activated protein kinase) and inhibition of mTORC1 (mammalian target of rapamycin complex 1) signaling pathway [11,12] leads to the activation of the ULK1/ULK2 (UNC51-like autophagy activating kinase 1/2) complex, which in turn activates the nucleation complex, including ATG13, RB1CC1 (RB1-inducible coiled-coil 1) and ATG101, which orchestrates the recruitment of further ATG proteins on the phagophore, promoting membrane elongation [13]. Nucleation complex phosphorylates AMBRA1 (autophagy and beclin 1 regulator 1) and BECN1 (beclin1), disrupting the inhibitory association with BCL2 (BCL2 apoptosis regulator) and allowing PI3KC3 (phosphatidylinositol 3-kinase catalytic subunit type 3) complex assembly [14]. The PI3KC3 complex, including the PI3KC3, PIK3R4 (PI3KC3 regulatory subunit 4), UVRAG (UV radiation resistance associated), AMBRA1 and BECN1, controls the membrane nucleation stage and initial phagophore formation [15]. PI3P (phosphatidylinositol 3-phosphate) that is generated by PI3K activity in the newly formed membranes, serves as a landing pad, known as omegasomes, for effector proteins such as ZFYVE1 (zinc finger FYVE-type containing 1) and WIPI2 (WD repeat domain phosphoinositide-interacting 2), to promote the formation of isolation membrane [16]. Elongation of the isolation membrane requires the involvement on two ubiquitin-like conjugation systems. In the first system, the protease ATG4 cleaves the precursor form of MAP1LC3A (microtubule associated protein 1 light chain 3 alpha), allowing ATG7 (E1 ubiquitin-activating enzyme) and ATG3 (E2 ubiquitin-conjugating enzyme) to catalyze the MAP1LC3A-I conjugation with phosphatidylethanolamine, to form MAP1LC3A-II [17,18]. At the same time, ATG12 is covalently conjugated to the ATG5 protein through the action of ATG7 and ATG10 (E2 ubiquitin-like enzyme) proteins [17,18]. Then, recruitment of the ATG16L1 protein (E3 ubiquitin-protein ligase) stabilizes the ATG12-ATG5 complex. ATG12-5-16L1 oligomers facilitates MAP1LC3A-II localization to the phagophore membrane, where it drives autophagosomal maturation. Once autophagosome maturation is finished, ATG4 catalyzes the reverse modification reaction of MAP1LC3A-II to MAP1LC3A-I [18]. Following completion and closure, the autophagosome ultimately undergoes fusion with a lysosome (Figure 1A). The fusion process is regulated by lysosomal membrane and cytoskeletal proteins. Finally, the fusion results in the exposure of autophagosomal cargo to the lysosomal acid hydrolases required for degradation and recycling [18]. Autophagy can be classified as non-selective or cargo-specific [19]. Selectivity of autophagy is ensured by two groups of autophagic cargo receptors, the ubiquitin-binding type, exemplified by SQSTM1 (sequestosome-1) [20] and CALCOCO2 (calcium binding and coiled-coil domain 2) [21], and the trans-membrane type, exemplified by BNIP3 (BCL2 interacting protein 3) and BNIP3L (BCL2 interacting protein 3 like) [22,23]. Although not essential for autophagosomal biogenesis itself, receptor proteins specifically bind the cargo material and the autophagosomal membrane, acting as a bridge that promotes cargo sequestration by the nascent autophagosome [24] (Figure 1B).