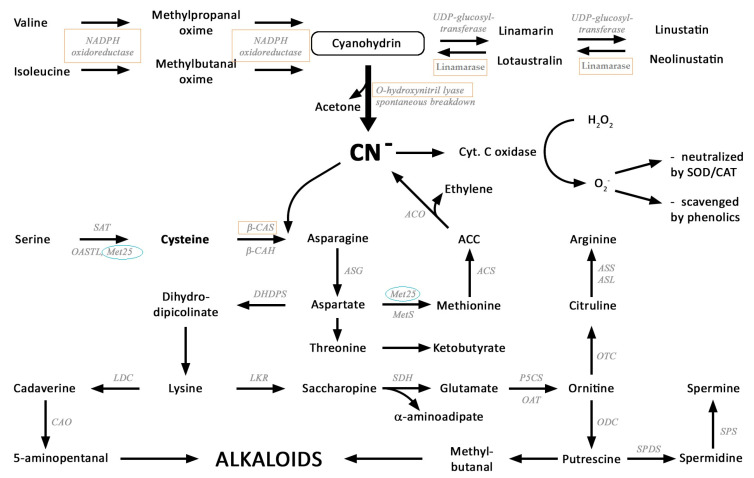
Figure 1.
Metabolism of cyanogenic glucosides. Key enzymes marked on the scheme: biosynthesis: NADPH oxidoreductase (Val-monooxygenase), UDP-glucosyl-transferase; catabolism: linamarase (β-D-glucosidase), O-hydroxynitryl lyase; CN− ions asymilation: β-CAS -β-cyanoalanine synthase, β-CAH-β-cyanoalanine hydratase, SAT-serine O-transacetylase, OASTL-O-acetylserine (thiol) lyase, ASG-asparaginase, MetS-methionine synthase, ACS-ACC synthase, ACO-ACC oxidase, DHDPS-dihydro-dipicolinate synthase, LDC-L-lysine decarboxylase, CAO-cadaverine oxydase, LKR-lysine-ketoglutarate reductase, SDH-sacchropine dehydrogenase, P5CS-pyrroline-5-carboxylate synthase, OAT-ornithine aminotransferase, ODC-ornithine decarboxylase, SPDS-spermidine synthase, SPS-spermine synthase, OTC-ornithine transcarbamylase, ASS-argininosuccinate synthetase, ASL-argininosuccinate lyase; ROS scavenging processes: CAT-catalase, SOD-superoxide dismutase. Exogenous gene Met25-O-acetylhomoserine-O-acetylserine (OAH-OAS) sulfhydrylase from Saccharomyces. cerevisiae. Genes whose expressions have been studied and described in this work are marked with red boxes. The blue ellipse marked the activity site of the Met25 yeast gene introduced into transgenic plants.