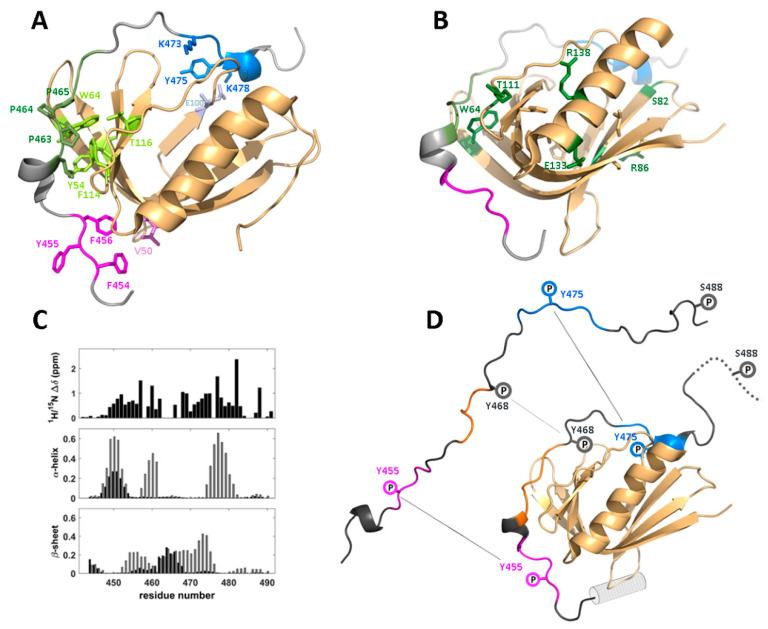
Figure 5.
Structural view of the interaction between the C-terminal domain of WIP and the WASp EVH1 domain. Structures are based on the complex between WIP residues 451–485 tethered to residues 26–147 of rat N-WASp (PDB ID: 2IFS [104]). Residue numbers are based on the analogous WASp sequence. (A) Structure of the WIP-N-WASp complex (adapted from [104]). N-WASp is shown in light orange, and three WIP epitopes are shown in magenta, green, and blue. Sidechain atoms of these epitopes and key N-WASp residues forming the binding interface are shown as sticks with a similar coloring scheme. (B) Distribution of WAS-causing mutations; residues that when mutated result in severe WAS, are highlighted with sidechains in stick representation. Buried mutation hotspot residues are colored in light-orange, and surface-exposed hotspot residues are colored in green and labeled. T111 represents the location of analogous N-WASp residue R601. (C) Chemical shift data indicating a binding-induced conformational change in WIP, including (top) HSQC perturbations along the sequence, (middle) predicted helical content for free (black) and bound (gray) WIP, (bottom) same as previous but for β-strand content. (D) Model of binding induced changes in residues 442–492 of WIP showing the additional helical motif binding to WASp (gray cylinder) and phospho-sites along the sequence.