Figure 3.
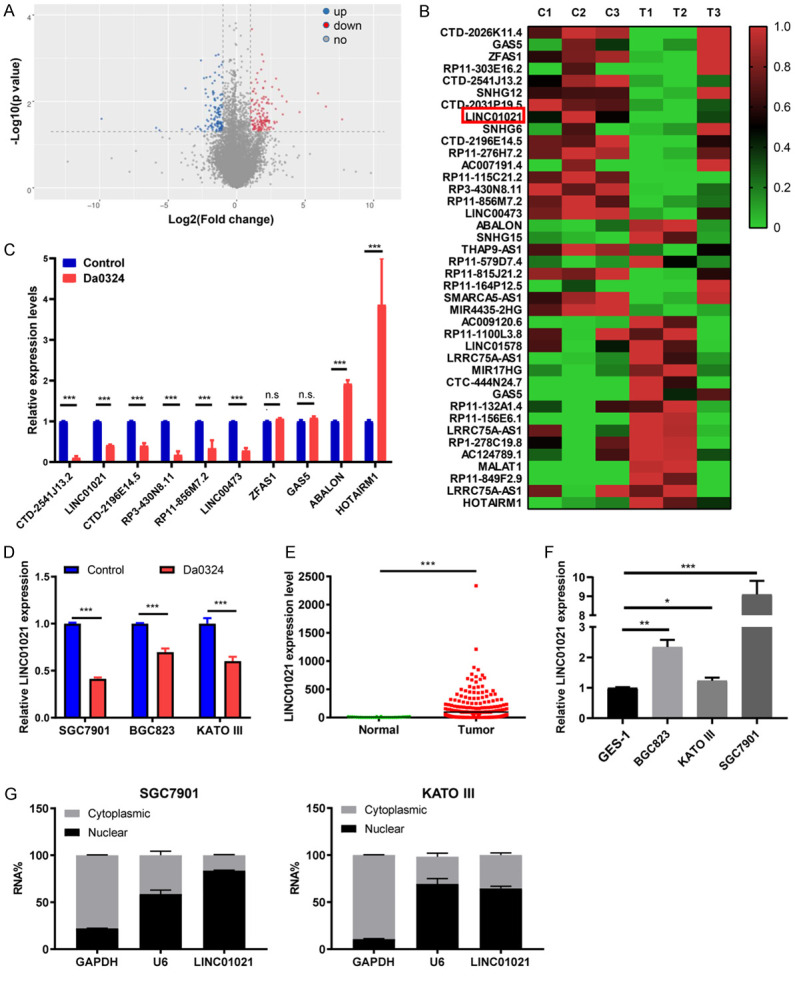
Long non-coding RNA LINC01021 was downregulated by Da0324 treatment in gastric cancer cells. A. Volcano plots of lncRNAs differentially expressed between the control and Da0324 treatment groups. SGC7901 cells were treated with DMSO (control) or 4 μM Da0324 for 48 h and high-throughput sequencing assay was performed. The X-axis represents log2 of fold changes. The Y-axis represents -log10 of P values. Red spots denote upregulated lncRNAs, blue spots denote downregulated lncRNAs. B. Heatmap of the top 40 lncRNAs most significantly differentially expressed in SGC7901 cells with Da0324 treatment. C. Ten differentially expressed lncRNAs regulated by Da0324 were validated by qRT-PCR. SGC7901 cells were treated with DMSO (control) or 4 μM Da0324 for 48 h. RNAs were isolated and converted to cDNA. Quantitative real-time PCR was performed to determine the expression level of lncRNAs. GAPDH was used as a housekeeping gene. All bars represent relative expression levels and data represent mean ± SD. ***, P < 0.001; n.s. means no significant difference. D. LINC01021 expression by qRT-PCR in BGC823, SGC7901 and KATO III cells treated with Da0324 or DMSO (control) for 48 h. All bars represent relative expression levels and data represent mean ± SD. ***, P < 0.001. E. TCGA data for the expression of LINC01021 in gastric cancer tissues (n = 375) and normal tissues (n = 32). F. LINC01021 expression by qRT-PCR in the normal gastric epithelial cell line GES-1 and in gastric cancer cell lines (BGC823, SGC7901, and KATO III). All bars represent relative expression levels and data represent mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. G. The subcellular location of LINC01021 in SGC7901 and KATO III cells was determined by qRT-PCR. GAPDH was used as a positive control for cytoplasmic RNA localization, U6 for nuclear RNAs. The data are shown as means ± SD. lncRNA, long non-coding RNA; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
