Abstract
Tumor endothelial cell marker 8 (TEM8) is a type I transmembrane protein, that has been widely studied in the areas of anthrax toxin infection and tumor angiogenesis. However, the role of TEM8 in the progression of epithelial ovarian cancer (EOC) remains unclear. In this study, we determined that TEM8 was highly expressed in ovarian cancer and associated with poor prognosis in EOC patients. In vitro experiments showed that TEM8 overexpression significantly promoted ovarian cancer proliferation. TEM8 overexpression also promoted the G0/G1 phase transition, migration, and invasion of ovarian cancer cells but suppressed apoptosis. Moreover, experimental verification confirmed that TEM8 overexpression increased the expression of Ki-67, cyclin D1, Bcl2/Bax, MMP2, MMP9, and VEGFA and the phosphorylation of Rac1/Cdc42, JNK, MEK, ERK, and STAT3 (Ser727). Subsequently, the addition of RAC1 (EHop-016) and MEK (PD98059) pathway inhibitors suppressed malignant behaviors in the TEM8 overexpression group, which robustly indicated that TEM8 activated Rac1/Cdc42/JNK and MEK/ERK/STAT3 signaling pathways. In addition, we also revealed that the transcription factor GATA2 bound to the TATTAGTTATCTTT site of the TEM8 promoter region and regulated its expression. In conclusion, our study may provide a new theoretical basis for TEM8 application as a clinical biomarker and potential target in EOC patients.
Keywords: TEM8, Rac1/Cdc42/JNK signaling pathway, MEK/ERK/STAT3 signaling pathway, GATA2, ovarian cancer
Introduction
Ovarian cancer (OC) is a lethal malignant tumor of the female reproductive system with the characteristics of insidious onset, chemotherapy resistance, and a low 5-year survival rate [1]. At present, CA125, human epididymis protein 4 (HE4), the risk of ovarian malignancy algorithm (ROMA) index, and image examination are used in the clinical diagnosis of OC. However, approximately 70% of patients are in advanced stages when diagnosed [2,3]. In addition, despite the great success of surgery and chemotherapy in the treatment of OC, recurrence and metastasis are still the main causes of death [4]. The overall 5-year survival rate of OC patients is approximately 47%, whereas the 5-year survival rate of stage III and IV OC patients is lower, 41% and 20%, respectively [5]. The invasion and metastasis of OC involves interactions between multiple genes and factors. Therefore, elucidating the molecular mechanism in epithelial ovarian cancer (EOC) can provide a theoretical basis for targeted therapy.
Tumor endothelial marker 8 (TEM8) is also known as anthrax toxin receptor 1 (ANTXR1) [6]. In 2000, St Croix identified 9 tumor endothelial markers (TEMs) using serial analysis of gene expression (SAGE), and only TEM8 was not detected in luteal formation, suggesting that TEM8 was associated with tumor-associated angiogenesis [7]. TEM8 is a type I transmembrane protein, and its extracellular domain (N-terminal) contains a vWA domain with a metal-ion-dependent adhesion site (MIDAS), which is involved in protein-protein interactions, such as α-integrin binding to collagen in the extracellular matrix (ECM). The intracellular domain (C-terminal) participates in protein phosphorylation and transmits corresponding cellular signals [8]. By constructing a chimera IL2R/TEM8-CD, i.e., the extracellular domain of IL-2R and the intracellular domain of TEM8, Werner E found that IL2R/TEM8 only bound actin not intermediate filaments and microtubules, and promoted cell migration and lamellipodia formation [9]. TEM8 is overexpressed in a variety of malignant tumors, and little is known about its role in EOC. Our previous study found that the expression of TEM8 was upregulated in the HE4 overexpression group compared to the control group, suggesting that TEM8 might play an important role in EOC progression [10].
In this study, we detected TEM8 expression in EOC using immunohistochemistry and determined the correlations between TEM8 expression and the clinicopathological parameters and prognosis of EOC patients. Specifically, we explored the effect of TEM8 overexpression and knockdown on the malignant behaviors of OC cells. Furthermore, we also detected the activated pathways and transcription factors that regulated TEM8 expression. The above findings provide a new theoretical basis for OC early diagnosis, follow-up monitoring and targeted therapy.
Materials and methods
Ethical statement
The study was approved by the Ethics Committee of Shengjing Hospital affiliated to China Medical University (#2019PS451K for human tissue). The study followed the Declaration of Helsinki and informed consents were obtained from all patients.
Tissue samples and clinical data
140 cases of paraffin-embedded pathological specimens were collected from the Department of Obstetrics and Gynecology in Shengjing Hospital affiliated to China Medical University from 2008 to 2014. All histological sections were diagnosed by pathologists. In total, 103, 13, 12, and 12 cases of malignant ovarian epithelial tumors (malignant group), borderline epithelial ovarian tumors (borderline group), benign ovarian epithelial tumors (benign group) and normal ovarian tissues (normal group) were collected, respectively. The median age of the patients was 53 years (19-79 years, malignant group), 43 years (19-84 years, borderline group), 51 years (28-78 years, benign group), and 45 years (32-76 years, normal group). There was no significant difference in age among the four groups (P > 0.05). In the malignant group, the distribution of pathological types was as follows: 73, 8, 15, and 7 cases of serous adenocarcinoma, mucinous adenocarcinoma, endometrioid adenocarcinoma and clear cell carcinoma, respectively. The pathological grading consisted of 26, 25, and 52 cases of well-differentiated, moderately-differentiated and poorly-differentiated, respectively. According to International Federation of Gynecology and Obstetrics (FIGO, 2009) classifications, there were 27, 15, 56, and 5 cases of stage I, stage II, stage III and stage IV, respectively. In addition, there were 20, 53, and 30 cases of lymph node metastasis, no metastasis and no lymphadenectomy, respectively. All cases were primary epithelial ovarian malignancies with complete clinical data. Radiotherapy, chemotherapy, and hormone therapy were not implemented before surgery.
Immunohistochemistry
Paraffin-embedded ovarian tissues were sliced at 5 μm continuously. The expression of TEM8 protein was detected using the immunohistochemical streptavidin-peroxidase (SP) method. Rabbit anti-TEM8 (1:100, Abcam, Shanghai, China) staining was carried out according to the SP kit (Maixin, Fujian, China) instructions. The brown-yellow particle of cell membranes and cytoplasm was considered a positive staining. The stain intensity was scored as follows: 0 score (no coloring), 1 scores (light yellow), 2 scores (brown yellow) and 3 scores (brown); The percentage of stained area was classified as follows: 0 score (< 5%), 1 scores (5-25%), 2 scores (26-50%), 3 scores (51-75%), and 4 scores (> 75%). The final score was obtained by multiplying the above two items: 0-2 scores (-), 3-4 scores (+), 5-8 scores (++), and 9-12 scores (+++). Finally, 0-4 scores were considered as the low expression groups and 5-12 scores were considerated as the high expression groups. Tissue stain images were judged independently by two senior pathologists who did not know the clinical data.
Cell culture and transfection
OVCAR3, SKOV3, and CAOV3 cells were purchased from American Type Culture Collection (Manassas, VA, USA), and A2780 cells were purchased from Jennio Biotech (Guangdong, China). All OC cell lines were cultured in RPMI 1640 containing 10% fetal bovine serum (FBS) at 37°C, 5% CO2, and saturated humidity. Cell lines in logarithmic growth phase were seeded in 6-well plates and transfected with TEM8 siRNA, GATA2 siRNA and negative control (all from Genepharma, Shanghai, China) according to the Lipofectamine 3000 instructions (Thermo Fisher Scientific Inc., US). The sequence of TEM8 siRNA is: sense: 5’-GCGGAUUUGACCUGUACUUTT-3’, antisense: 5’-AAGUACAGGUCAAAUCCGCTT-3’. The sequence of GATA2 siRNA is: sense: 5’-GGUGGACGUCUUCUUCAAUTT-3’, antisense: 5’-AUUGAAGAAGACGUCCACCTT-3’. The sequence of its negative control is: sense: 5’-UUCUCCGAACGUGUCACGUTT-3’, antisense: 5’-ACGUGACACGUUCGGAGAATT. Stable TEM8 overexpression (TEM8-H) and control (TEM8-H-Mock) cell lines were established according to the GeneChem lentiviral gene transfection instructions and selected with puromycin (2-4 μg/ml, Solarbio, Beijing, China).
Real-time quantitative PCR
Total RNA was extracted using the TRIzol (Takara Bio, Inc., Shiga, Japan) method. The purity and quantitation were determined using an ultraviolet spectrophotometer. Total RNA (1 μg) was reverse transcribed into cDNA using the Takara 047A kit (Takara Bio, Inc., Shiga, Japan), and SYBR fluorescent dye-labeled real-time PCR was performed using a 7500 Fast Real-Time PCR system. TEM8 primer sequence: Forward: 5’-AAAGTGGCCAACGGTAGACG-3’, Reverse: 5’-TGCTCCGGCATCTTGACTCT-3’. GAPDH primer sequence: Forward: 5’-ACAACTTTGGTATCGTGGAAGG-3’, Reverse: 5’-GCCATCACGCCACAGTTTC-3’. Data were analyzed using the -ΔCT method.
MTT assay
Cells were trypsinized and diluted to a concentration of 1×104/ml and 200 μl per well was seeded in a 96-well plate. After 6 h, cells had adhered to the plate, so this was defined as time 0. Then, 20 μl MTT (5 mg/ml, Solarbio, Beijing, China) was added and incubated for 4 h, the solution was aspirated, and 150 μl DMSO was added. After shaking for 10 min, the optical density (OD) at 490 nm was measured at 0, 24, 48, 72 and 96 h.
Scratch assay
Cells in the logarithmic growth phase were seeded in 6-well plates at approximately 5×105 cells/ml/well. When the cell monolayer was covered with medium, scratches were performed using a 200-μl pipette tip. The cells were washed two to three times with PBS to remove cell debris and cultured with serum-free medium. The scratch width was measured at 0 and 24 h under the microscope.
Transwell assay
RPMI 1640 medium (500 μl) with 20% FBS was added to the lower chamber of the gelled transwell. Serum-free cell suspension (2.5×105 cells/ml, 200 μl) was added to the upper chamber and incubated at 37°C, 5% CO2 for 24 or 48 h. Then, chambers were fixed with 4% paraformaldehyde for 30 min, washed with PBS, stained with 0.1% crystal violet, and the number of cells that passed through the Matrigel was counted under the microscope.
Cell apoptosis assay
Cell apoptosis rates were detected using annexin-V-FITC/PI (Dojindo Molecular Technologies, Kumamoto, Japan) and annexin V-PE/7AAD (BD Biosciences, New York, USA) kits. A single cell suspension was prepared by trypsinization, followed by centrifugation at 1000 rpm for 5 min. The supernatant was removed and 300 μl 1× annexin V binding buffer (1×106 cells/mL) was added. Then, 5 μl annexin V-FITC binding mixture (annexin V-PE binding mixture) and 5 μl PI dye (7AAD dye) were added to the cell suspension. The cells were incubated in the dark at room temperature for 15 min, and apoptosis was detected using flow cytometry BD FACSDiva Software (BD Biosciences, New York, USA).
Cell cycle assay
A single cell suspension was prepared by trypsinization and centrifuged at 1000 rpm for 5 min. The supernatant was removed, 500 μl of precooled 70% ethanol was added, and the cells were fixed overnight at 4°C. The next day, the fixative was washed away with PBS, 500 μl of PI/RNase A staining solution (KeyGen Biotech, Nanjing, China) was added, and the mixture was shielded from light for 30 min at room temperature. The ratio of G0/G1, S, and G2/M phase cells was detected using flow cytometry BD FACSDiva Software (BD Biosciences, New York, USA).
Western blotting
Cells were rinsed and centrifuged at 2,000 rpm for 5 min, and the total protein was extracted using the RIPA method and quantified using the BCA method. Proteins were separated using 10% SDS-PAGE, transferred to PVDF membranes and blocked using 5% BSA at room temperature for 2 h. Primary antibodies were incubated at 4°C overnight (dilution ratio of 1:1000): rabbit anti-TEM8 (Abcam, Cambridge, UK), rabbit anti-GATA2 (Abcam, Cambridge, UK), rabbit anti-Rac1 (Abcam, Cambridge, UK), rabbit anti-Cdc42 (Wanleibio, Shenyang, China), rabbit anti-p-Rac1/cdc42 (Cell Signaling Technology, California, USA), rabbit anti-JNK (Cell Signaling Technology, California, USA), mouse anti-p-JNK (Cell Signaling Technology, California, USA), rabbit anti-STAT3 (Wanleibio, Shenyang, China), rabbit anti-MEK (Santa Cruz, USA), rabbit anti-p-MEK (Cell Signaling Technology, California, USA), rabbit anti-ERK (Cell Signaling Technology, California, USA), rabbit anti-p-ERK (Cell Signaling Technology, California, USA), rabbit anti-p-STAT3 (Ser727) (Wanleibio, Shenyang, China), anti-Ki-67 (Wanleibio, Shenyang, China), anti-cyclin D1 (Cell Signaling Technology, California, USA), rabbit anti-MMP2 (Proteintech, Wuhan, China), anti-rabbit MMP9 (Proteintech, Wuhan, China), mouse anti-Bcl2 (Proteintech, Wuhan, China), rabbit anti-Bax (Proteintech, Wuhan, China), rabbit anti-VEGFA (Abcam, Cambridge, UK), mouse anti-GAPDH (1:2000, ZSGB-BIO Technology Co., Ltd., Beijing, China). The next day, the membranes were washed three times using 1×TBST, the secondary antibody was added and incubated for 2 h at room temperature, and the membranes were washed three times using 1×TBST. The protein bands were detected using ECL chemiluminescence (Millipore, USA).
Pathway inhibitors
RAC1 (EHop-016, Selleck Chemicals, USA) and MAPK (PD98059, Selleck Chemicals, USA) inhibitors were dissolved in DMSO, and cells were treated with doses of 2 μM and 20 μM for 48 h, respectively. The cells were collected for further experiments.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed according to the Simple ChIP Enzymatic Chromatin IP Kit (#9004, Cell Signaling Technology, California, USA) instructions. Briefly, 1% formaldehyde fixation, nuclear preparation and chromatin digestion, chromatin immunoprecipitation, elution and reverse cross-linking, and purification of DNA were performed. Finally, 2% lysate was used as an input control, the remaining lysate was immunoprecipitated with a rabbit IgG or GATA2 antibody, and the immunoprecipitated DNA was amplified using RT-qPCR. The binding site primer sequence: Forward: 5’-GGTCAAGTAAGTCTGGGAAAC-3’, Reverse: 5’-AACTACCCAGCTAAGCCATT-3’.
Bioinformatic analysis
Coexpressed genes (|Spearman’s Correlation| ≥ 0.5) in OC were downloaded from the cBioPortal website. DAVID (version 6.8) was employed to analyze the terms including GO terms (biological process/cellular component/molecular function, BP/CC/MF) and KEGG pathways. ggplot2. The R package was used for visualization. Normalized gene expression RNAseq data of OC were downloaded from the UCSC Xena database (https://xenabrowser.net/). Samples were ranked from low to high according to TEM8 expression, and divided into four equal parts. The first 25% of the samples were considered as the low expression group, and the last 25% were considered the high expression group. The datasets for c2.cp.kegg.v6.2.symbols.gmt and c2.cp.biocarta.v6.2.symbols.gmt were downloaded from the MsigDB database of the GSEA website and then analyzed using GSEA version 3.0 software. The JASPAR database was used to predict the binding site of transcription factors to the TEM8 promoter region. The StarBase_v3.0 database was used to analyze the mRNA correlation.
Statistical analysis
All data were analyzed using SPSS 21.0 (IBM Corporation, Armonk, NY, USA) software and graphed using GraphPad Prism 8.0 software. All data were presented as the mean ± SD. The chi-squared test and Student’s t-test were used to compare the differences between two groups, and analysis of variance (ANOVA) was used to compare differences among more than two groups. Kaplan-Meier survival curves were used for prognostic analysis, and univariate and multivariate Cox regression were used to analyze risk factors for prognosis. Two-sided P < 0.05 was considered statistically significant. (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Results
TEM8 expression in different ovarian tissues and its clinical significance
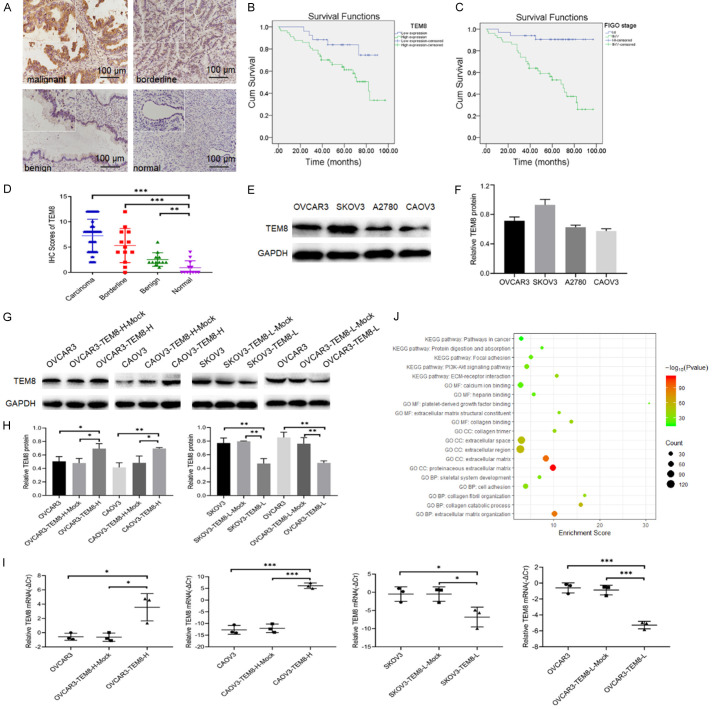
TEM8 expression was located in the cell membrane and cytoplasm (Figure 1A). Its positive rate and high positive rate in the malignant group were 90.29% and 69.90%, respectively, which were significantly higher than in the benign (33.3% and 8.33%) and normal (16.7% and 0%) groups (P < 0.05 for all; Table 1). However, the positive (P = 0.161) and high positive rate (P = 0.343) of TEM8 was not statistically different between the malignant and borderline groups. Immunohistochemical scores of the malignant and borderline groups were 7.26 ± 3.25 and 5.31 ± 3.38, significantly higher than the normal group (0.92 ± 1.38; P < 0.001 for all; Figure 1D).
Figure 1.
High TEM8 expression correlates with poor prognosis of EOC patients and its function prediction using DAVID: (A) TEM8 expression in various ovarian tissues (×200, scale bar = 100 μm; upper left ×400): malignant ovarian epithelial tumors (n = 103), borderline epithelial ovarian tumors (n = 13), benign ovarian epithelial tumors (n = 12), normal ovarian tissues (n = 12). (B, C) Influence of TEM8 expression and FIGO stage on the overall survival of ovarian cancer patients. (D) Immunostaining score of TEM8 in malignant, borderline, benign, and normal ovarian tissues. (E, F) Representative images and quantitation of the western blot showed that the protein expression of TEM8 in four OC cell lines (n = 3). GAPDH was used as an internal control. (G, H) Representative images and quantitation of the western blot showed that the protein expression of TEM8 in the TEM8 overexpression/knockdown groups (n = 3). GAPDH was used as an internal control. (I) The relative TEM8 mRNA expression in the TEM8 overexpression/knockdown groups (n = 3). (J) The bubble plot showed top5 GO (BP/CC/MF) and KEGG pathway of coexpressed genes of TEM8. Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Table 1.
TEM8 expression in different ovarian tissues
| Groups | Cases | Low | High | Positive rate (%) | High Positive rate (%) | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| - | + | ++ | +++ | ||||
| Malignant | 103 | 10 | 21 | 42 | 30 | 90.29a,b | 69.90c,d |
| Borderline | 13 | 3 | 3 | 5 | 2 | 76.92e,f | 53.85g,h |
| Benign | 12 | 8 | 3 | 1 | 0 | 33.33 | 8.33 |
| Normal | 12 | 10 | 2 | 0 | 0 | 16.7 | 0 |
malignant vs. benign (***, P < 0.001);
malignant vs. normal (***, P < 0.001);
malignant vs. benign (***, P < 0.001);
malignant vs. normal (***, P < 0.001);
borderline vs. benign (*, P = 0.028);
borderline vs. normal (**, P = 0.003);
borderline vs. benign (*, P = 0.03);
borderline vs. normal (**, P = 0.005).
In total, 103 EOC cases were included in the study. The chi-square test showed that a high positive rate of TEM8 expression was significantly correlated with FIGO stage (P = 0.001) and lymph node metastasis (P = 0.004), but showed no correlation with age (P = 0.424), pathological type (P = 0.216), or differentiation (P = 0.562; Table 2). Kaplan-Meier analysis showed that high positive TEM8 expression (P < 0.05; Figure 1B) and stage III-IV (P < 0.05; Figure 1C) was significantly associated with poor survival of patients with EOC.
Table 2.
Relationship between TEM8 expression and clinicopathological parameters of 103 patients with EOC
| Parameters | Cases | TEM8 expression | Positive rate (%) | P | High Positive rate (%) | P | |||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| - | + | ++ | +++ | ||||||
| Age (median 53 years) | |||||||||
| <53 | 47 | 6 | 10 | 23 | 8 | 87.23 | 0.506 | 65.96 | 0.424 |
| ≥53 | 56 | 4 | 11 | 19 | 22 | 92.86 | 73.21 | ||
| pathologic | |||||||||
| Serous | 73 | 5 | 18 | 31 | 19 | 93.15 | 0.146 | 68.49 | 0.216 |
| Mucinous | 8 | 2 | 2 | 1 | 3 | 75.0 | 50.0 | ||
| Endometrioid | 15 | 3 | 0 | 8 | 4 | 80.0 | 80.0 | ||
| Clear cell | 7 | 0 | 1 | 2 | 4 | 100.0 | 85.71 | ||
| FIGO stage | |||||||||
| I-II | 42 | 6 | 14 | 13 | 9 | 85.71 | 0.310 | 52.38 | 0.001** |
| III-IV | 61 | 4 | 7 | 29 | 21 | 93.44 | 81.97 | ||
| Differentiation | |||||||||
| Well-moderate | 51 | 6 | 8 | 26 | 11 | 88.24 | 0.526 | 72.55 | 0.562 |
| Poor | 52 | 4 | 13 | 16 | 19 | 92.31 | 67.31 | ||
| LN metastasis | |||||||||
| No | 53 | 7 | 14 | 15 | 17 | 86.79 | 0.179 | 60.38 | 0.004** |
| Yes | 20 | 0 | 1 | 12 | 7 | 100.0 | 95.0 | ||
| unknown | 30 | 3 | 6 | 15 | 6 | 90.0 | 70.0 | ||
Abbreviations: FIGO, International Federation of Gynecology and Obstetrics. LN, lymph node.
P < 0.01.
Then, we analyzed 73 cases of EOC, except for 30 cases without lymphadenectomy. Univariate Cox regression analysis showed that TEM8 expression, FIGO stage, and lymph node metastasis were independent risk factors for EOC (P < 0.05 for all). However, multivariate Cox regression analysis showed that only FIGO stage was a risk factor for EOC (P < 0.05; Table 3).
Table 3.
Univariate and multivariate Cox regression analysis of clinical pathological Parameters in 73 patients with EOC
| Parameters | groups | Univariate analysis | P | Multivariate analysis | P | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| HR | 95% CI | HR | 95% CI | ||||
| Age | <53 | 1.627 | (0.672-3.941) | 0.281 | |||
| ≥53 | |||||||
| FIGO stage | I-II | 9.892 | (2.295-42.634) | 0.002** | 7.224 | (1.548-33.719) | 0.012* |
| III-IV | |||||||
| Differentiation | Well-moderate | 1.368 | (0.573-3.269) | 0.480 | |||
| poor | |||||||
| Lymph node metastasis | No | 3.161 | (1.320-7.568) | 0.010* | 1.303 | (0.523-3.242) | 0.570 |
| Yes | |||||||
| TEM8 expression | Low | 4.466 | (1.039-19.20) | 0.044* | 2.308 | (0.514-10.368) | 0.275 |
| High | |||||||
P < 0.05;
P < 0.01.
High TEM8 expression promoted OC cell proliferation but reduced apoptosis
The expression of TEM8 protein in OVCAR3, SKOV3, A2780, and CAOV3 OC cell lines was detected using western blotting (Figure 1E, 1F). The results showed that TEM8 expression was highest expression in SKOV3 and OVCAR3 cells, followed by A2780 and CAOV3 cells. Therefore, we constructed stable TEM8 overexpression cell lines (OVCAR3-TEM8-H and CAOV3-TEM8-H) and TEM8 knockdown cell lines (SKOV3-TEM8-L and OVCAR3-TEM8-L). QRT-PCR and western blotting confirmed that the TEM8 mRNA and protein levels were increased/decreased in the TEM8 overexpression/knockdown groups (Figure 1G-I).
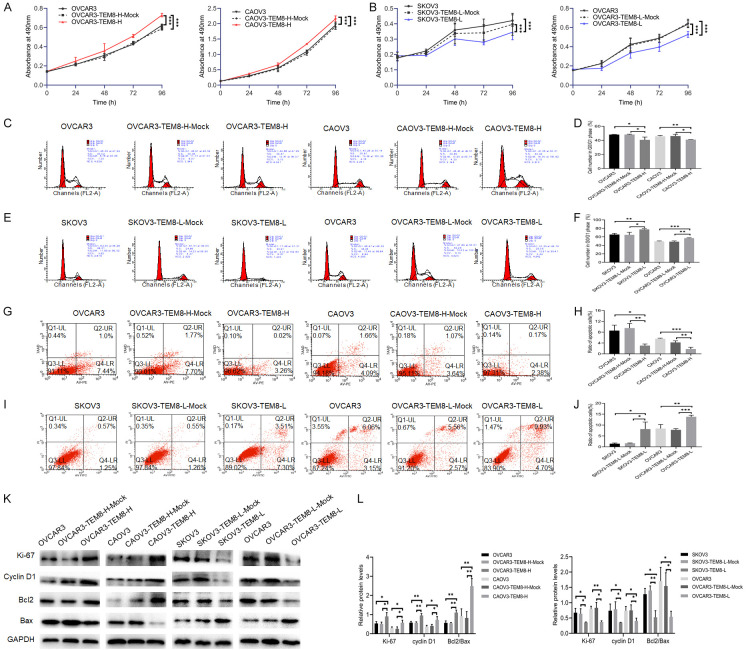
The MTT assay showed that cell proliferation was significantly increased (P < 0.05 for all; Figure 2A). Cell cycle and cell apoptosis assay showed the G0/G1 phase population (P < 0.05 for all; Figure 2C, 2D) and apoptosis rate (P < 0.05 for all; Figure 2G, 2H) were significantly decreased in the TEM8 overexpression groups compared to the negative control (NC) groups. While cell proliferation was decreased (P < 0.05 for all; Figure 2B), the G0/G1 phase population (P < 0.05 for all; Figure 2E, 2F) and apoptosis rate (P < 0.05 for all; Figure 3I, 3J) were significantly increased in the TEM8 knockdown groups compared to the NC groups. Western blotting showed that the ratio of Ki-67, cyclin D1, and Bcl2/Bax was increased in the TEM8 overexpression groups, and these results were reversed in the TEM8 knockdown groups (P < 0.05 for all; Figure 3K, 3L). The above findings indicated that high TEM8 expression promoted proliferation and G0/G1 transition, but inhibited the apoptosis of OC cells.
Figure 2.
TEM8 promoted proliferation but reduced apoptosis in ovarian cancer cells. A, B. MTT assay showed that TEM8 overexpression increased cell proliferation of ovarian cancer cells, however, these trends were reversed in the TEM8 knockdown groups (n = 9). C-F. Cell cycle assay showed that TEM8 overexpression promoted G0/G1 phase transition of ovarian cancer cells, however, these trends were reversed in the TEM8 knockdown groups (n = 3). G-J. Cell apoptosis assay showed that TEM8 overexpression reduced apoptosis rate of ovarian cancer cells, however, these trends were reversed in the TEM8 knockdown groups (n = 3). K, L. Representative images and quantitation of the western blot showed that the protein expression of Ki-67, cyclin D1, Bcl2, and Bax in the TEM8 overexpression/knockdown groups (n = 3). GAPDH was used as an internal control. Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
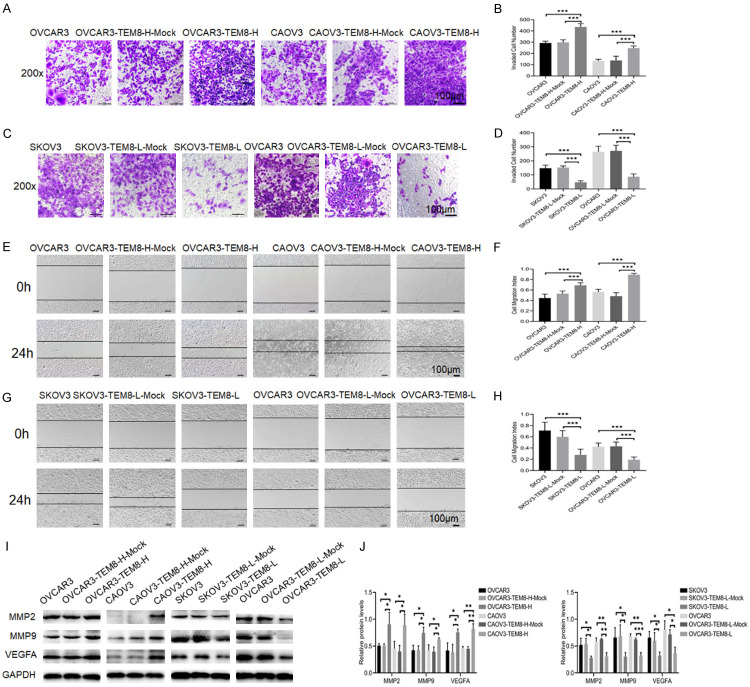
Figure 3.
TEM8 promoted invasion and migration in ovarian cancer cells. A-D. Transwell assay showed that TEM8 overexpression promoted invasion of ovarian cancer cells, however, TEM8 knockdown reversed this effect (n = 9; ×200, scale bar = 100 μm). E-H. Scratch assay showed that TEM8 overexpression promoted migration of ovarian cancer cells, however, TEM8 knockdown reversed this effect (n = 9; ×100, scale bar = 100 μm). I, J. Representative images and quantitation of the western blot showed that the protein expression of MMP2, MMP9, and VEGFA in the TEM8 overexpression/knockdown groups (n = 3). GAPDH was used as an internal control. Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
High TEM8 expression promoted the migration and invasion of OC cells
Coexpressed genes refer to genes that are correlated with each other in a particular biological process. Here, we screened coexpressed genes using cBioPortal and analyzed the GO terms (BP/CC/MF) and KEGG pathways associated with these gene sets using the DAVID database (version 6.8). A total of 29 BPs, 11 CCs, 11 MFs, and 8 KEGG pathways were enriched (P < 0.0001 for all), such as extracellular matrix organization, proteinaceous extracellular matrix, collagen binding, ECM-receptor interaction, etc., and the bubbles of the top5 GO terms (BP/CC/MF) and KEGG pathways were plotted using ggplot2.R language package (Figure 1J).
We performed scratch and Transwell tests to investigate the effects of TEM8 on migration and invasion of OC cells. Compared with those in the NC groups, the invasion (P < 0.05; Figure 3A, 3B) and migration (P < 0.05; Figure 3E, 3F) abilities were significantly increased in the OVCAR3-TEM8-H and CAOV3-TEM8-H groups. The invasion (P < 0.05; Figure 3C, 3D) and migration (P < 0.05; Figure 3G, 3H) abilities were significantly decreased in the TEM8 knockdown groups. Western blot results showed that the expression levels of MMP2, MMP9, and VEGFA were upregulated in the TEM8 overexpression groups, and these trends were reversed in the TEM8 knockdown groups (P < 0.05 for all; Figure 3I, 3J). The results indicated that TEM8 participated in the invasion and migration of OC cells.
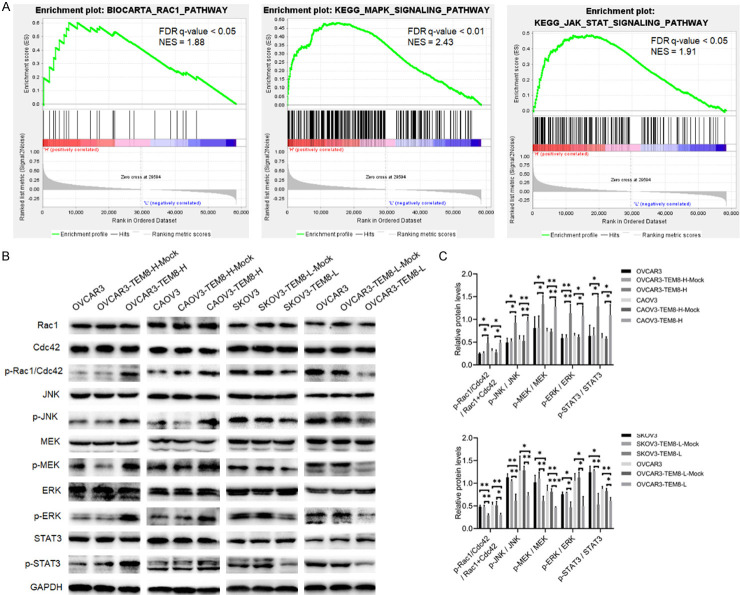
TEM8 activated Rac1/Cdc42/JNK and MEK/ERK/STAT3 signaling pathways
To investigate TEM8-mediated molecular pathways in OC, we conducted pathway enrichment using GSEA. The results showed that samples with high TEM8 expression were enriched for RAC1, MAPK, and JAK-STAT signaling pathways (FDR q-value < 0.05 for all; Figure 4A). Western blot analysis showed that p-Rac1/Cdc42, p-JNK p-MEK, p-ERK, and p-STAT3 (Ser727) were significantly increased in the OVCAR3-TEM8-H and CAOV3-TEM8-H groups compared to the NC groups. Conversely, the expression of these proteins was decreased in the SKOV3-TEM8-L and OVCAR3-TEM8-L groups (P < 0.05 for all; Figure 4B, 4C).
Figure 4.
TEM8 activated the Rac1/Cdc42/JNK and MEK/ERK/STAT3 signaling pathway. A. GSEA enrichment analysis suggested that TEM8 was associated with RAC1, MAPK, and JAK-STAT signaling pathways. B, C. Representative images and quantitation of the western blot showed that the protein expression of Rac1, Cdc42, p-Rac1/Cdc42, JNK, p-JNK, MEK, p-MEK, ERK, p-ERK, STAT3, and p-STAT3 in the TEM8 overexpression/knockdown groups (n = 3). GAPDH was used as an internal control. Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
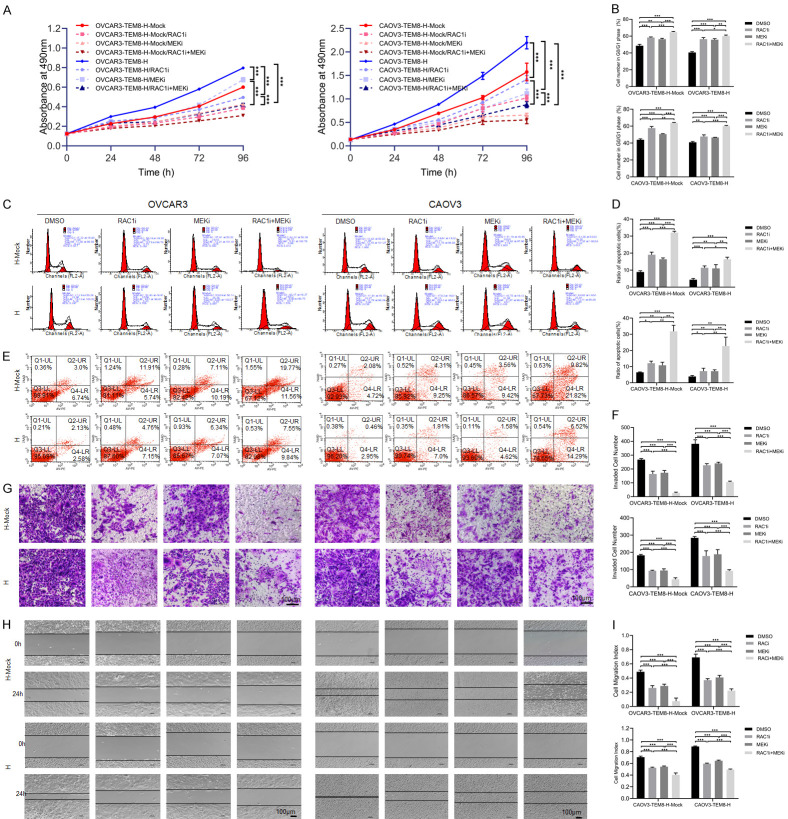
Next, we added RAC1 (EHop-016) and MEK (PD98059) pathway inhibitors to the TEM8 overexpression groups. The results showed that cell proliferation, migration and invasion were suppressed, whereas the G0/G1 phase and apoptosis rates were increased in the EHop-016 or PD98059 group compared to the NC groups. The combination of EHop-016 and PD98059 significantly attenuated cell proliferation (P < 0.05; Figure 5A, 5B), invasion (P < 0.05; Figure 5F, 5G), and migration (P < 0.05; Figure 5H, 5I) but increased the G0/G1 phase population (P < 0.05; Figure 5B, 5D) and apoptosis rate (P < 0.05; Figure 5C, 5E) compared with that observed with a single pathway inhibitor. These results strongly indicated that TEM8 overexpression promoted OC progression through the activation of the Rac1/Cdc42/JNK and MEK/ERK/STAT3 signaling pathways.
Figure 5.
Inhibition of Rac1/Cdc42/JNK and MEK/ERK/STAT3 pathway reduced TEM8-mediated proliferation, migration, invasion and apoptosis inhibition. A. MTT assay showed that addition of Rac1 (EHop-016) and MEK (PD98059) pathway inhibitors reduced cell proliferation in TEM8 overexpression groups (n = 9). B, C. Cell cycle assay showed that addition of Rac1 (EHop-016) and MEK (PD98059) pathway inhibitors inhibited G0/G1 phase transition in TEM8 overexpression groups (n = 3). D, E. Cell apoptosis assay showed that addition of Rac1 (EHop-016) and MEK (PD98059) pathway inhibitors increased apoptosis rate in TEM8 overexpression groups (n = 3). F, G. Transwell assay showed that addition of Rac1 (EHop-016) and MEK (PD98059) pathway inhibitors inhibited cell invasion in TEM8 overexpression groups (n = 9; ×200, scale bar = 100 μm). H, I. Scratch assay showed that addition of Rac1 (EHop-016) and MEK (PD98059) pathway inhibitors inhibited cell migration in TEM8 overexpression groups (n = 9; ×100, scale bar = 100 μm). Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
GATA2 regulated TEM8 expression at the transcriptional level
When exploring transcription factors regulating TEM8 expression, we identified the transcription factor GATA2 using the JASPAR database. The ChIP assay showed that GATA2 bound to the TEM8 upstream TATTAGTTATCTTT sequence (Figure 6A-C). Western blot results showed that the protein expression level of TEM8 was decreased in the GATA2 knockdown groups (P < 0.05; Figure 6E, 6F). We further verified the correlation between GATA2 mRNA and TEM8 mRNA expression in 379 cases of OC using starBase version 3.0. The results showed that there was a positive correlation between them (P < 0.05, r = 0.301; Figure 6D). These results indicated that GATA2 regulated the expression of TEM8 at the transcriptional level.
Figure 6.
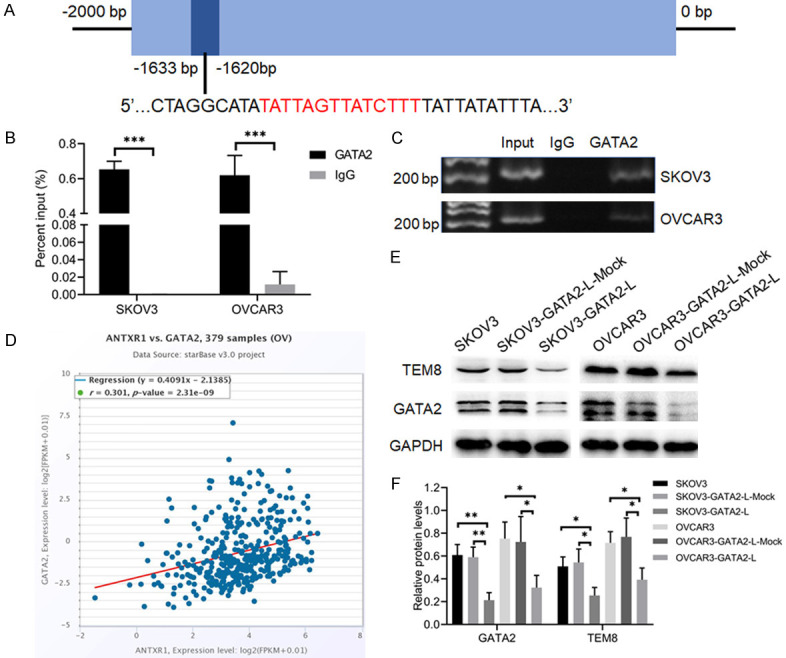
Transcription factor GATA2 regulated TEM8 expression. A. Prediction of GATA2 binding sites in the TEM8 promoter region. B, C. ChIP assay and agarose gel electrophoresis showed that GATA2 could bind the promoter region of TEM8 (n = 3). D. Correlation between TEM8 mRNA and GATA2 mRNA in 379 ovarian samples using starBase. E, F. Representative images and quantitation of the western blot showed that the protein expression of TEM8 and GATA2 in the GATA2 knockdown groups (n = 3). GAPDH was used as an internal control. Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
The increase in tumor invasion and metastasis is a landmark of increased malignancy of the tumor. OC has the characteristics of insidious onset, prone to pelvic and abdominal cavity implantation, and chemotherapy resistance, which poses a huge challenge for clinical diagnosis and treatment [11,12]. Therefore, exploring the molecular mechanism of OC invasion and metastasis provides new ideas for finding new molecular markers and targeted therapy.
TEM8 is a type I transmembrane protein that is expressed in normal tissues such as the lung, small intestine, and skin [13]. Numerous studies show that TEM8 is highly expressed in tumor tissues such as colon cancer, breast cancer, and osteosarcoma and participates in tumor angiogenesis, proliferation and adhesion and maintains tumor stem cell characteristics [14-16]. Our previous study demonstrated that the overexpression of the ovarian tumor marker HE4 upregulated TEM8 expression, suggesting that TEM8 was associated with EOC progression [10]. In this study, we found that TEM8 was highly expressed in the malignant group, and the TEM8 expression was mainly located in the cell membrane and cytoplasm. A high TEM8 expression rate was associated with FIGO stage and lymph node metastasis (P < 0.05). Kaplan-Meier results showed that high TEM8 expression rate was associated with poor prognosis in EOC patients. In addition, univariate Cox regression analysid demonstrated that TEM8 expression, FIGO stage and lymph node metastasis were independent risk factors for EOC (P < 0.05). The above results indicated that TEM8 participated in the progression of EOC, and might be one of the biomarkers for early detection and prognosis evaluation.
Cellular movement is a key step in the breakthrough of tumor cells into the basement membrane and distant metastasis and is related to the remodeling of the cytoskeleton and ECM [17]. Similar to the role of the integrin family, the extracellular domain of TEM8 mediates cell movement by interacting with ECM components such as collagen I and IV, and its intracellular domain interacts with cytoskeletal proteins [8,9]. A previous study demonstrated that TEM8 coimmunoprecipitated with α-smooth muscle actin (α-SMA), which resulted in a conformational change of TEM8 and promoted its binding to ECM components [18]. Here, we discovered that TEM8 was closely related to ECM remodeling, cell adhesion, and cell migration in OC using cBioPortal and DAVID. Our results showed that TEM8 overexpression increased the migration and invasion of OC cells, which was in line with the bioinformatic findings. In addition, TEM8 overexpression promoted OC cell proliferation and G0/G1 transition but inhibited apoptosis. To clarify the underlying mechanism, GSEA enrichment analysis showed that TEM8 was associated with the RAC1, MAPK, and JAK-STAT signaling pathways. MAPK belongs to a Ser/Thr protein kinase family, mainly including the ERK, JNK, p38MAPK and ERK5 subfamilies [19]. Studies have reported that RAC1 activates the JNK signaling pathway and MEK/ERK activates the STAT3 signaling pathway [20,21]. Therefore, we speculated that TEM8 might activate the Rac1/Cdc42/JNK and MEK/ERK/STAT3 signaling pathways in OC cells. In this study, we verified the above signaling pathway proteins in the TEM8 overexpression and knockdown groups. The results suggested that the phosphorylation levels of Rac1/Cdc42, JNK, MEK, ERK, and STAT3 were increased in the TEM8 overexpression groups, and were decreased in the TEM8 knockdown groups. RAC1 and MEK pathway inhibitor addition decreased the proliferation, migration and invasion ability of OC cells, and increased the apoptosis rate in the TEM8 overexpression group. These results indicated that TEM8 promoted the malignant biological behavior of OC cells by activating the Rac1/Cdc42/JNK and MEK/ERK/STAT3 signaling pathways.
Rac1 and Cdc42 are members of the Rho GTPase family, and these GTPase have two states: GTP (activated) and GDP (non-activated), and the activated form serves as a “molecular switch” in proliferation, angiogenesis, and cytoskeletal remodeling [22-24]. Studies have shown that Rac1 was highly expressed in OC tissues and was associated with poor prognosis [25]. The allosteric inhibitor R-ketorolac inhibited the invasion and metastasis of OC by inhibiting the activity of Rac1 and Cdc42 [26]. GTP-Rac1/Cdc42 can regulate actin cytoskeleton remodeling and activate JNK signaling pathway, thereby promoting cell proliferation and migration [20,27-29]. It was reported that basic fibroblast growth factor (bFGF) activated the PI3K/Akt-Rac1-FAK-JNK pathway, thus promoting melanoma migration [30]. MEK/ERK is one of the MAPK signaling pathways involved in the regulation of cell proliferation, invasion and migration [31,32]. p-ERK promotes transcription expression and protein activation of matrix metalloproteinases MMP2 and MMP9, contributing to ECM degradation, cell migration, and invasion [33]. Studies found that TEM8 regulated the expression of p21, p27, and cyclin D1 by activating the ERK1/2 signaling pathway and promoting osteosarcoma proliferation [16]. Signal transduction and transcriptional activator (STAT3) is a member of the STAT protein family, which mainly receives signals from the membrane receptors IL-6R, IL-21R, and IL-23R, and plays an important role in regulating cell proliferation and apoptosis [34,35]. p-STAT3 (Ser727) can be activated by MAPK and the nonreceptor tyrosine kinase c-src and participates in the transcription of downstream genes such as Bcl-2, Bcl-xl, and VEGFA [21,36-38]. Fukumoto T et al. showed that myeloid leukemia-1 (Mcl-1L) inhibited UVB-induced melanoma cell apoptosis by activating the MEK/ERK/STAT3 pathway [39]. Our results demonstrated that the expression levels of Ki-67, cyclin D1, Bcl2/Bax, MMP2, MMP9, and VEGFA were significantly increased in the TEM8 overexpression group compared to the NC group. The above study indicated that TEM8 upregulated the expression of Ki-67, cyclin D1, Bcl2/Bax, MMP2, MMP9, and VEGFA by activating the Rac1/Cdc42/JNK and MEK/ERK/STAT3 signaling pathways, thus promoting the malignant biological behavior of OC cells.
GATA2 is a transcription factor with two zinc finger domains: C-ZnF is responsible for binding to specific DNA sites, whereas N-ZnF is responsible for interacting with various other nuclear proteins. GATA2 can bind to a nucleic acid sequence (AGATCTTA or AGATAA) at the promoter and enhancer sites and transcriptionally activate or inhibit the expression of targeted genes [40]. The initial research on GATA2 was mainly focused on the hematopoietic system, which played an important role in regulating the proliferation and differentiation of hematopoietic stem cells and hematopoietic progenitor cells, and its abnormal expression was associated with acute myeloid leukemia and aplastic anemia [41,42]. The role of GATA2 in solid tumors is gradually being recognized. It was found that GATA2 overexpression was related to the progression of prostate cancer and breast cancer [43,44]. GATA2 served as a coactivator for androgen receptor (AR) and synergistically regulated gene expression, thus promoting the progression of prostate cancer [43]. One study suggested that GATA2 could be used as a novel biomarker for OC through multiomics analysis [45]. Our study demonstrated for the first time that GATA2 bound to the TATTAGTTATCTTT region of the TEM8 promoter and regulated TEM8 expression. TEM8 protein was significantly decreased in GATA2 knockdown groups. The results also showed that GATA2 mRNA was positively correlated with TEM8 mRNA using the starBase_v3.0 database. Therefore, we speculated that GATA2 regulated the expression of TEM8 at the transcriptional level.
Currently, bevacizumab application has achieved progress by regulating antitumor angiogenesis, but it also has side effects on physiological angiogenesis [46]. TEM8 is highly expressed in various tumors and tumor-associated endothelial cells, but not in neovascularization during corpus luteum formation [47], which indicates that TEM8 can be used as a targeted gene to selectively inhibit tumor progression. In addition, as anthrax toxin receptors, TEM8 and ANTXR2 can mediate the virulence of anthrax toxin (PA) into cells, but ANTXR2 is widely expressed in both normal and tumor tissues [48]. Therefore, the application of engineered PA to TEM8-overexpressing tumor tissue could become a hotspot in the field of tumor biotherapy [49].
In summary, this study demonstrated that TEM8 was highly expressed in EOC and associated with a poor prognosis. High TEM8 expression promoted OC cell proliferation, G0/G1 transition, migration, and invasion but reduced apoptosis by activating the MEK/ERK/STAT3 and Rac1/Cdc42/JNK signaling pathways. The expression of TEM8 was regulated by the transcription factor GATA2. These findings have important implications for TEM8 application as a clinical biomarker and potential target to inhibit the progression of OC.
Acknowledgements
The authors would like to acknowledge all the members in Dr. Lin B’s laboratory for help with this study. This work was supported by the National Natural Science Foundation of China (grants 81672590 and 81472437) and Shengjing Freelance Research Program (grant 201804).
Disclosure of conflict of interest
None.
References
- 1.Ottevanger PB. Ovarian cancer stem cells more questions than answers. Semin Cancer Biol. 2017;44:67–71. doi: 10.1016/j.semcancer.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Sandri MT, Bottari F, Franchi D, Boveri S, Candiani M, Ronzoni S, Peiretti M, Radice D, Passerini R, Sideri M. Comparison of HE4, CA125 and ROMA algorithm in women with a pelvic mass: correlation with pathological outcome. Gynecol Oncol. 2013;128:233–238. doi: 10.1016/j.ygyno.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 3.Korsunsky I, Parameswaran J, Shapira I, Lovecchio J, Menzin A, Whyte J, Dos Santos L, Liang S, Bhuiya T, Keogh M, Khalili H, Pond C, Liew A, Shih A, Gregersen PK, Lee AT. Two microRNA signatures for malignancy and immune infiltration predict overall survival in advanced epithelial ovarian cancer. J Investig Med. 2017;65:1068–1076. doi: 10.1136/jim-2017-000457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narod S. Can advanced-stage ovarian cancer be cured? Nat Rev Clin Oncol. 2016;13:255–261. doi: 10.1038/nrclinonc.2015.224. [DOI] [PubMed] [Google Scholar]
- 5.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu J, Faundez V, Werner E. Endosomal recycling regulates Anthrax Toxin Receptor 1/Tumor Endothelial Marker 8-dependent cell spreading. Exp Cell Res. 2010;316:1946–1957. doi: 10.1016/j.yexcr.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 8.Hotchkiss KA, Basile CM, Spring SC, Bonuccelli G, Lisanti MP, Terman BI. TEM8 expression stimulates endothelial cell adhesion and migration by regulating cell-matrix interactions on collagen. Exp Cell Res. 2005;305:133–144. doi: 10.1016/j.yexcr.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 9.Werner E, Kowalczyk AP, Faundez V. Anthrax toxin receptor 1/tumor endothelium marker 8 mediates cell spreading by coupling extracellular ligands to the actin cytoskeleton. J Biol Chem. 2006;281:23227–23236. doi: 10.1074/jbc.M603676200. [DOI] [PubMed] [Google Scholar]
- 10.Zhu L, Guo Q, Jin S, Feng H, Zhuang H, Liu C, Tan M, Liu J, Li X, Lin B. Analysis of the gene expression profile in response to human epididymis protein 4 in epithelial ovarian cancer cells. Oncol Rep. 2016;36:1592–1604. doi: 10.3892/or.2016.4926. [DOI] [PubMed] [Google Scholar]
- 11.Tan DS, Agarwal R, Kaye SB. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol. 2006;7:925–934. doi: 10.1016/S1470-2045(06)70939-1. [DOI] [PubMed] [Google Scholar]
- 12.Kipps E, Tan DS, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat Rev Cancer. 2013;13:273–282. doi: 10.1038/nrc3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonuccelli G, Sotgia F, Frank PG, Williams TM, de Almeida CJ, Tanowitz HB, Scherer PE, Hotchkiss KA, Terman BI, Rollman B, Alileche A, Brojatsch J, Lisanti MP. ATR/TEM8 is highly expressed in epithelial cells lining Bacillus anthracis’ three sites of entry: implications for the pathogenesis of anthrax infection. Am J Physiol Cell Physiol. 2005;288:C1402–1410. doi: 10.1152/ajpcell.00582.2004. [DOI] [PubMed] [Google Scholar]
- 14.Rmali KA, Watkins G, Harrison G, Parr C, Puntis MC, Jiang WG. Tumour endothelial marker 8 (TEM-8) in human colon cancer and its association with tumour progression. Eur J Surg Oncol. 2004;30:948–953. doi: 10.1016/j.ejso.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 15.Chen D, Bhat-Nakshatri P, Goswami C, Badve S, Nakshatri H. ANTXR1, a stem cell-enriched functional biomarker, connects collagen signaling to cancer stem-like cells and metastasis in breast cancer. Cancer Res. 2013;73:5821–5833. doi: 10.1158/0008-5472.CAN-13-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao C, Wang Z, Huang L, Bai L, Wang Y, Liang Y, Dou C, Wang L. Down-regulation of tumor endothelial marker 8 suppresses cell proliferation mediated by ERK1/2 activity. Sci Rep. 2016;6:23419. doi: 10.1038/srep23419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins C, Nelson WJ. Running with neighbors: coordinating cell migration and cell-cell adhesion. Curr Opin Cell Biol. 2015;36:62–70. doi: 10.1016/j.ceb.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang MY, Chaudhary A, Seaman S, Dunty J, Stevens J, Elzarrad MK, Frankel AE, St Croix B. The cell surface structure of tumor endothelial marker 8 (TEM8) is regulated by the actin cytoskeleton. Biochim Biophys Acta. 2011;1813:39–49. doi: 10.1016/j.bbamcr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coso OA, Chiariello M, Yu JC, Teramoto H, Crespo P, Xu N, Miki T, Gutkind JS. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 21.Tkach M, Rosemblit C, Rivas MA, Proietti CJ, Diaz Flaque MC, Mercogliano MF, Beguelin W, Maronna E, Guzman P, Gercovich FG, Deza EG, Elizalde PV, Schillaci R. p42/p44 MAPK-mediated Stat3Ser727 phosphorylation is required for progestin-induced full activation of Stat3 and breast cancer growth. Endocr Relat Cancer. 2013;20:197–212. doi: 10.1530/ERC-12-0194. [DOI] [PubMed] [Google Scholar]
- 22.Hudson LG, Gillette JM, Kang H, Rivera MR, Wandinger-Ness A. Ovarian tumor microenvironment signaling: convergence on the Rac1 GTPase. Cancers (Basel) 2018;10:358. doi: 10.3390/cancers10100358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choudhari R, Minero VG, Menotti M, Pulito R, Brakebusch C, Compagno M, Voena C, Ambrogio C, Chiarle R. Redundant and nonredundant roles for Cdc42 and Rac1 in lymphomas developed in NPM-ALK transgenic mice. Blood. 2016;127:1297–1306. doi: 10.1182/blood-2015-11-683052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz J, Franke K, Frick M, Schumacher S. Different roles of the small GTPases Rac1, Cdc42, and RhoG in CALEB/NGC-induced dendritic tree complexity. J Neurochem. 2016;139:26–39. doi: 10.1111/jnc.13735. [DOI] [PubMed] [Google Scholar]
- 25.Leng R, Liao G, Wang H, Kuang J, Tang L. Rac1 expression in epithelial ovarian cancer: effect on cell EMT and clinical outcome. Med Oncol. 2015;32:329. doi: 10.1007/s12032-014-0329-5. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, Kenney SR, Muller CY, Adams S, Rutledge T, Romero E, Murray-Krezan C, Prekeris R, Sklar LA, Hudson LG, Wandinger-Ness A. R-ketorolac targets Cdc42 and Rac1 and alters ovarian cancer cell behaviors critical for invasion and metastasis. Mol Cancer Ther. 2015;14:2215–2227. doi: 10.1158/1535-7163.MCT-15-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acevedo A, Gonzalez-Billault C. Crosstalk between Rac1-mediated actin regulation and ROS production. Free Radic Biol Med. 2018;116:101–113. doi: 10.1016/j.freeradbiomed.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Snider JL, Allison C, Bellaire BH, Ferrero RL, Cardelli JA. The beta1 integrin activates JNK independent of CagA, and JNK activation is required for Helicobacter pylori CagA+-induced motility of gastric cancer cells. J Biol Chem. 2008;283:13952–13963. doi: 10.1074/jbc.M800289200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 30.Shi H, Lin B, Huang Y, Wu J, Zhang H, Lin C, Wang Z, Zhu J, Zhao Y, Fu X, Lou Z, Li X, Xiao J. Basic fibroblast growth factor promotes melanocyte migration via activating PI3K/Akt-Rac1-FAK-JNK and ERK signaling pathways. IUBMB Life. 2016;68:735–747. doi: 10.1002/iub.1531. [DOI] [PubMed] [Google Scholar]
- 31.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35:600–604. doi: 10.3109/10799893.2015.1030412. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Bao C, Ma Z, Xu B, Ying X, Liu X, Zhang X. Perfluorooctanoic acid stimulates ovarian cancer cell migration, invasion via ERK/NF-kappaB/MMP-2/-9 pathway. Toxicol Lett. 2018;294:44–50. doi: 10.1016/j.toxlet.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Shen Y, Wang S, Shen Q, Zhou X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 2018;415:117–128. doi: 10.1016/j.canlet.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C, Zhang Z, Chen L, Lee HW, Ayrapetov MK, Zhao TC, Hao Y, Gao J, Yang C, Mehta GU, Zhuang Z, Zhang X, Hu G, Chin YE. Acetylation within the N- and C-terminal domains of src regulates distinct roles of STAT3-mediated tumorigenesis. Cancer Res. 2018;78:2825–2838. doi: 10.1158/0008-5472.CAN-17-2314. [DOI] [PubMed] [Google Scholar]
- 37.Kim SM, Lee JH, Sethi G, Kim C, Baek SH, Nam D, Chung WS, Kim SH, Shim BS, Ahn KS. Bergamottin, a natural furanocoumarin obtained from grapefruit juice induces chemosensitization and apoptosis through the inhibition of STAT3 signaling pathway in tumor cells. Cancer Lett. 2014;354:153–163. doi: 10.1016/j.canlet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 38.Pan L, Xiao H, Liao R, Chen Q, Peng C, Zhang Y, Mu T, Wu Z. Fatty acid binding protein 5 promotes tumor angiogenesis and activates the IL6/STAT3/VEGFA pathway in hepatocellular carcinoma. Biomed Pharmacother. 2018;106:68–76. doi: 10.1016/j.biopha.2018.06.040. [DOI] [PubMed] [Google Scholar]
- 39.Fukumoto T, Iwasaki T, Okada T, Hashimoto T, Moon Y, Sakaguchi M, Fukami Y, Nishigori C, Oka M. High expression of Mcl-1L via the MEK-ERK-phospho-STAT3 (Ser727) pathway protects melanocytes and melanoma from UVB-induced apoptosis. Genes Cells. 2016;21:185–199. doi: 10.1111/gtc.12330. [DOI] [PubMed] [Google Scholar]
- 40.Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tien FM, Hou HA, Tsai CH, Tang JL, Chiu YC, Chen CY, Kuo YY, Tseng MH, Peng YL, Liu MC, Liu CW, Liao XW, Lin LI, Lin CT, Wu SJ, Ko BS, Hsu SC, Huang SY, Yao M, Chou WC, Tien HF. GATA2 zinc finger 1 mutations are associated with distinct clinico-biological features and outcomes different from GATA2 zinc finger 2 mutations in adult acute myeloid leukemia. Blood Cancer J. 2018;8:87. doi: 10.1038/s41408-018-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganapathi KA, Townsley DM, Hsu AP, Arthur DC, Zerbe CS, Cuellar-Rodriguez J, Hickstein DD, Rosenzweig SD, Braylan RC, Young NS, Holland SM, Calvo KR. GATA2 deficiency-associated bone marrow disorder differs from idiopathic aplastic anemia. Blood. 2015;125:56–70. doi: 10.1182/blood-2014-06-580340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao JC, Fong KW, Jin HJ, Yang YA, Kim J, Yu J. FOXA1 acts upstream of GATA2 and AR in hormonal regulation of gene expression. Oncogene. 2016;35:4335–4344. doi: 10.1038/onc.2015.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, He X, Ngeow J, Eng C. GATA2 negatively regulates PTEN by preventing nuclear translocation of androgen receptor and by androgen-independent suppression of PTEN transcription in breast cancer. Hum Mol Genet. 2012;21:569–576. doi: 10.1093/hmg/ddr491. [DOI] [PubMed] [Google Scholar]
- 45.Gov E, Kori M, Arga KY. Multiomics analysis of tumor microenvironment reveals Gata2 and miRNA-124-3p as potential novel biomarkers in ovarian cancer. OMICS. 2017;21:603–615. doi: 10.1089/omi.2017.0115. [DOI] [PubMed] [Google Scholar]
- 46.Bellati F, Napoletano C, Gasparri ML, Ruscito I, Marchetti C, Pignata S, Tomao F, Benedetti Panici P, Nuti M. Current knowledge and open issues regarding bevacizumab in gynaecological neoplasms. Crit Rev Oncol Hematol. 2012;83:35–46. doi: 10.1016/j.critrevonc.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Seaman S, Stevens J, Yang MY, Logsdon D, Graff-Cherry C, St Croix B. Genes that distinguish physiological and pathological angiogenesis. Cancer Cell. 2007;11:539–554. doi: 10.1016/j.ccr.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci U S A. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen KH, Liu S, Bankston LA, Liddington RC, Leppla SH. Selection of anthrax toxin protective antigen variants that discriminate between the cellular receptors TEM8 and CMG2 and achieve targeting of tumor cells. J Biol Chem. 2007;282:9834–9845. doi: 10.1074/jbc.M611142200. [DOI] [PMC free article] [PubMed] [Google Scholar]