Figure 1.
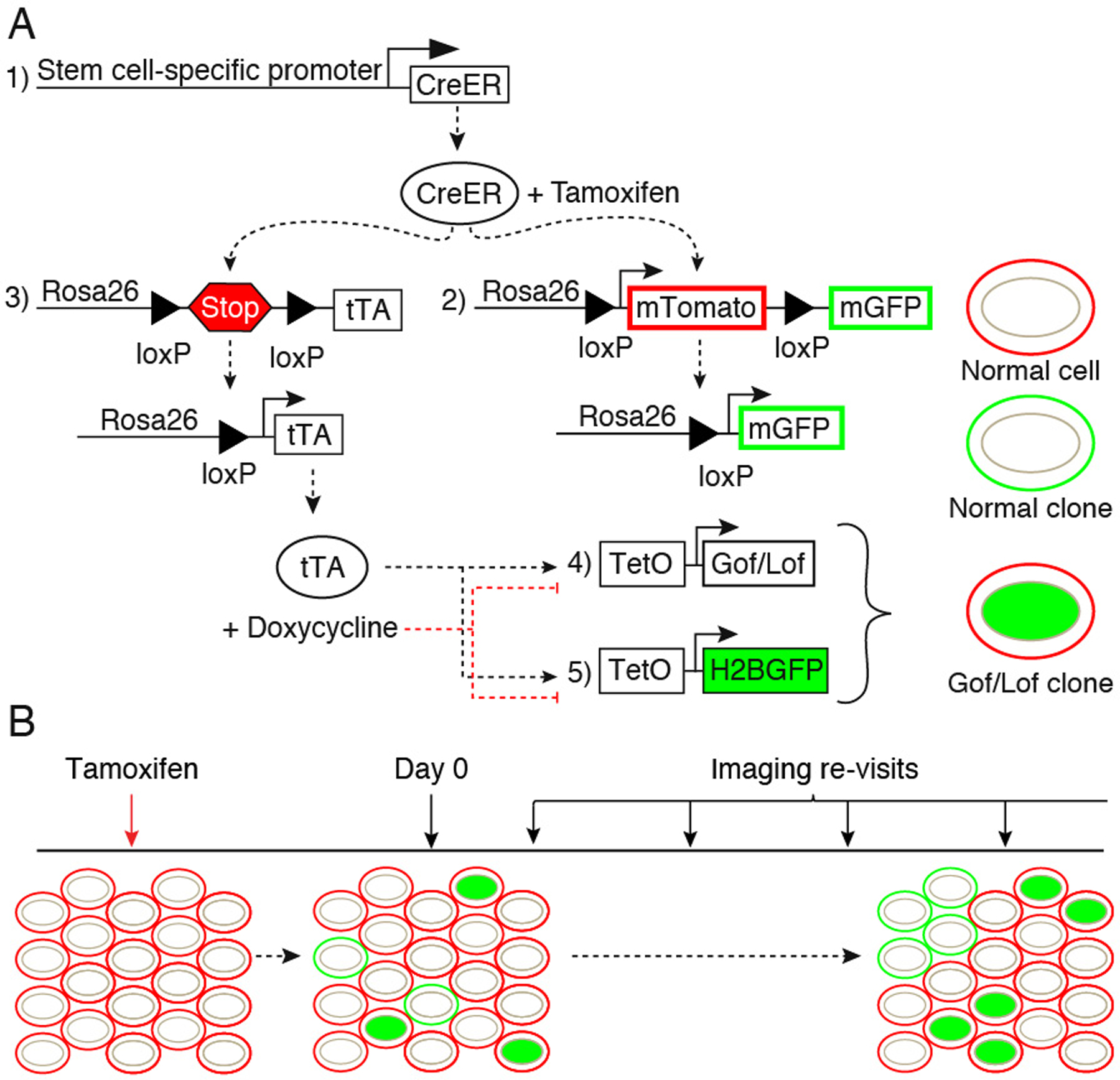
Genetic system and experimental timeline. To generate single genetically altered and normal clones, 5 alleles are used in our system. A stem cell-specific driver to control expression of an inducible Cre recombinase (1). Addition of Tamoxifen will enable the Cre-mediated excision of loxP-flanked sequences in two alleles: a reporter allele (2), in this case mT/mG, in which membrane-localized EGFP (mG) replaces the excised membrane-localized tdTomato (mT) in single clones, and the Tet transactivator allele (3), in this case LSL-tTA, where excision of a STOP cassette allows tTA expression. Expression of tTA in single clones will then enable activation of the TetO alleles: A gain- or loss-of-function (Gof/Lof) allele to alter a gene/pathway of interest (4) and a second reporter allele (5), in this case TetO-H2BGFP used to label the single “Lof/Gof clones” with nuclear EGFP. In this example, where the TetOFF system is used, doxycycline treatment will prevent binding of tTA to the TetO sequences, thus silencing TetO-driven gene expression (A). Before Tamoxifen treatment, all cells are uniformly labeled with membrane tdTomato (“normal cells”). After induction of Cre-mediated recombination, on the first imaging session taking place at “Day 0”, single clones labeled either with membrane EGFP instead of red (“normal clones”), or carrying a nuclear EGFP (“Gof/Lof clones”) are identified. Subsequent imaging sessions allow live tracking of clonal dynamics through time. The frequency of imaging revisits should be empirically determined according to experimental needs (B).
