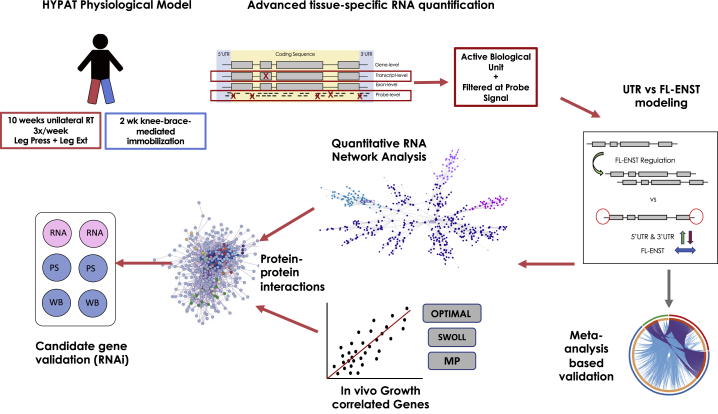
Figure 1.
Experimental Workflow and Analysis Strategy
We used a paired unilateral loading (10 weeks of progressive RT) and unloading (UL; 2 weeks of UL) model in combination with genome-wide transcriptomic analysis (Timmons et al., 2018) to study differential expression of gene UTRs and protein coding regions. Probes were subjected to extensive filtering before downstream analysis (see STAR Methods). Significance analysis of microarrays implemented in the R programming environment (SAMR) was used to detect significantly regulated genes (Tusher et al., 2001), which were then used as an input list for quantitative network analysis using the MEGENA package for R (Song and Zhang, 2015). We also determined which genes played a role in regulating dynamic lean tissue growth in independent cohorts (total n = 100). Highly co-regulated genes and growth-correlated gene lists were used as input in metascape.org and https://www.networkanalyst.ca to characterize protein-protein interaction networks and drug signatures.