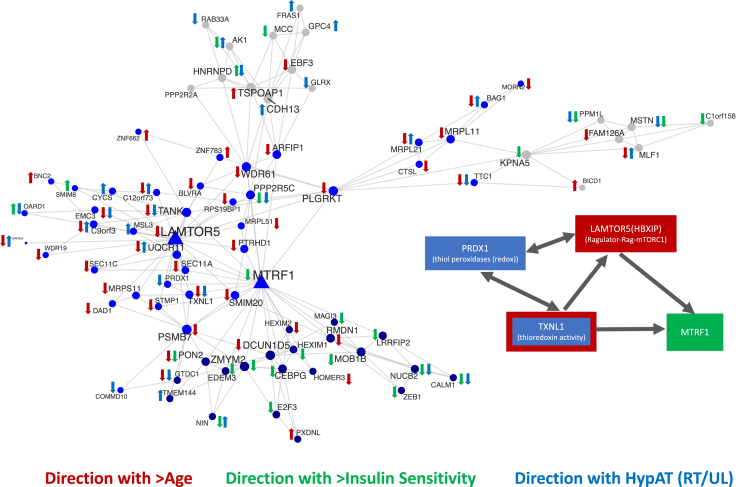
Figure 6.
Quantitative Network Analysis Unveils Potentially Important Gene Interactions across Potentially Related Physiological Conditions
An example network of gene interactions across different, yet interrelated physiological conditions, constructed using HypAt-, age-, and insulin sensitivity-regulated transcripts (FDR < 5%) as input into Megena (FDR < 1% Spearman correlation; p < 0.01 for module significance, p < 0.01 for network connectivity, and 10,000 permutations for calculating FDR and connectivity p values). This example network is centered on LAMTOR5, a gene encoding a subunit of the pentameric Ragulator complex involved in mTORC1 activation. From these interactome networks, relationships between genes can be discovered and subsequently studied in model systems. Triangle symbols represent ”hub genes,” whereas circles represent non-hub network members. Node colors represent different subnetwork clusters, and node size is proportional to node degree.