FIGURE 7.
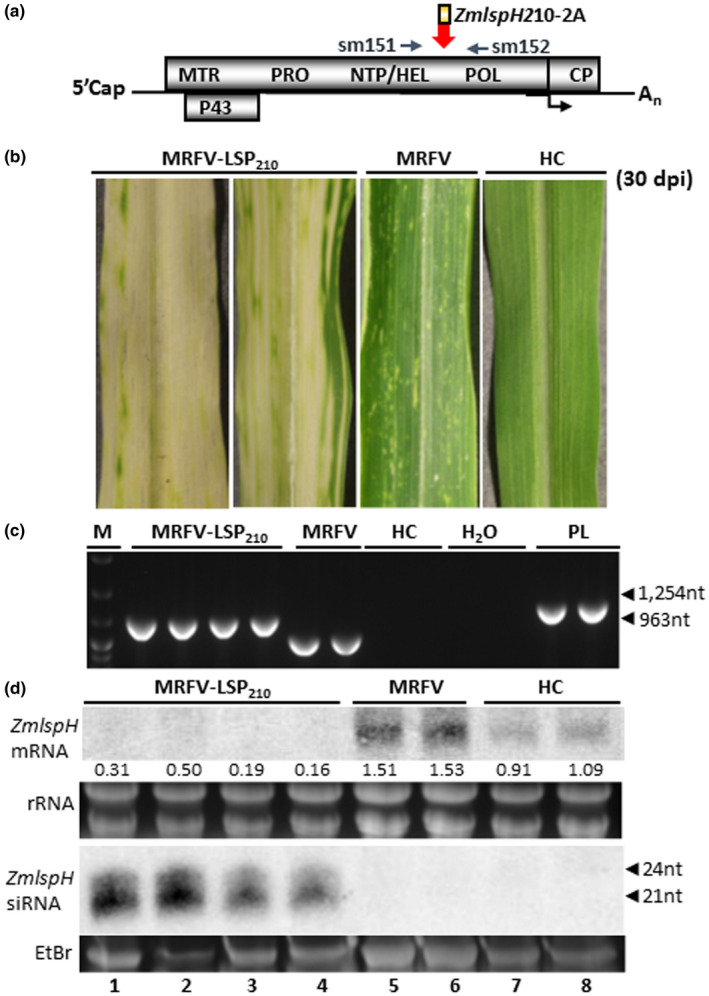
Efficacy of MRFV VIGS system in silencing ZmlspH gene. (a) Molecular assays for MRFV‐LSP210 used primers sm151 and 152 for RT‐PCR detection, amplifying 1,254 bp in MRFV‐LSP210 and 963bp in MRFV‐WT. (b) MRFV symptoms and yellow (albino) VIGS phenotype induced by MRFV‐LSP210 at 30 dpi, compared to plants inoculated with MRFV‐WT or noninoculated (HC). (c) RT‐PCR analysis of MRFV‐LSP210 and MRFV‐WT accumulation in systemic leaves 30 dpi. Noninoculated plants (HC) were included as negative controls; MRFV‐LSP210 plasmid (PL) served as a PCR‐positive control. M: 100 bp DNA marker. (d) Northern blot analysis of VIGS induced by MRFV‐LSP210. RNA blots were hybridized with LSP‐specific RNA probes to detect LSP mRNAs or antisense siRNAs. rRNA: EtBr‐stained ribosomal RNA used as loading control. For small RNA gel, the major low molecular weight RNA species was EtBr‐stained for loading control prior to bloating of gel onto membrane. Levels of LSP mRNA and siRNAs from MRFV‐LSP210‐infected plants (lanes 1–4) compared to MRFV‐infected (lanes 5–6) and healthy plants (lanes 7–8) are shown, with positions of 21‐ to 24‐nucleotide RNA size markers indicated by arrow heads. The relative levels of LSP mRNA were determined as described for PDS in Figure 3
