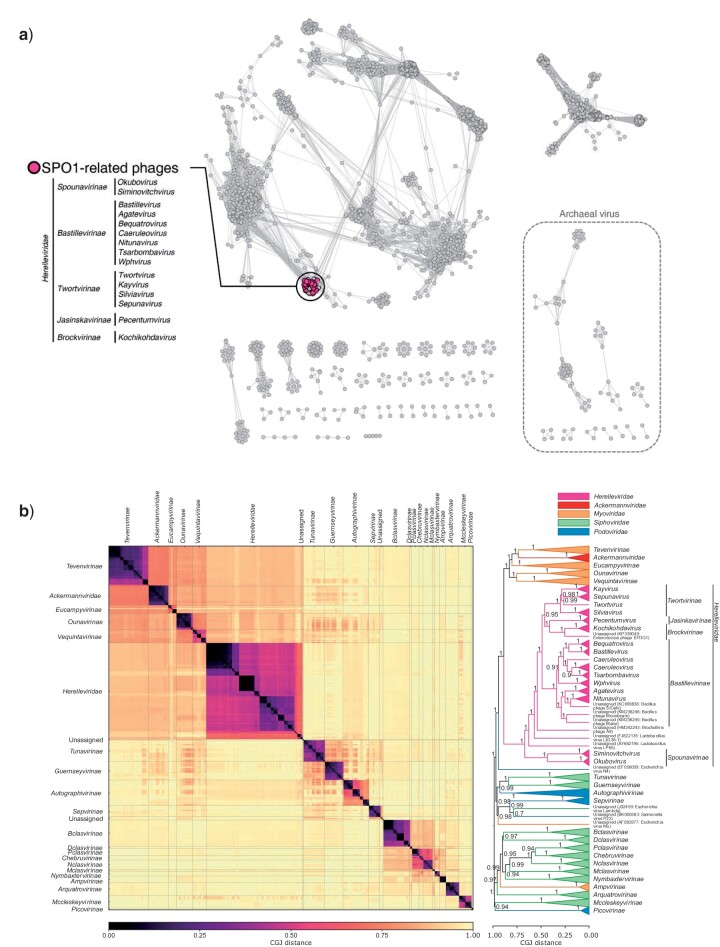
Figure 1.
a) Network representation of predicted protein content similarity of dsDNA viruses generated with vConTACT v2.0. Viruses are represented as circles (nodes) connected with each other (edges) based on a significant number of shared protein clusters, with more similar genomes displayed closer together on the network. The genomes belonging to the new family Herelleviridae are indicated with a circle. Genomes previously assigned to the subfamily Spounavirinae are indicated in pink. b) Clustering of dsDNA bacteriophages that possess subfamily assignments in the order Caudovirales generated with GRAViTy, darker colors in the heatmap represent higher degrees of similarity between genomes. The phages are clustered using UPGMA into a dendrogram, showing bootstrap values (100 pseudoreplicates) on each branch.