Figure S6. Detection of histidine-rich glycoprotein peptides in human serum by MS/MS analysis.
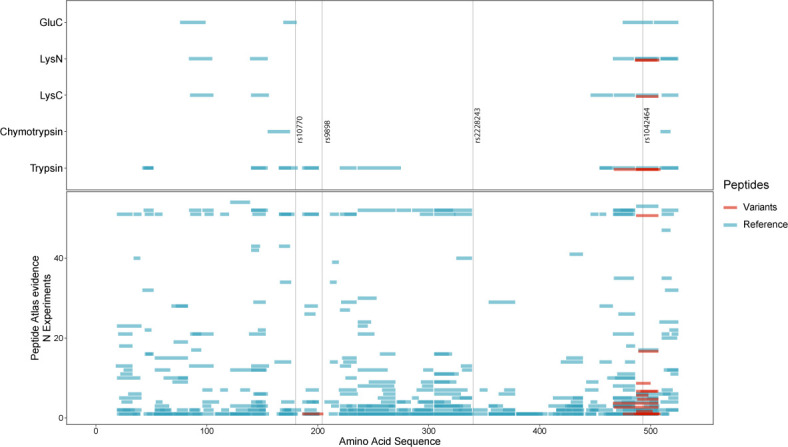
Peptides from histidine-rich glycoprotein detected in serum samples using a bottom-up proteomic experimental workflow and liquid chromatography tandem mass spectrometry for read-out. Top panel: five different enzymes (y-axis; GluC, LysN, LysC, chymotrypsin, and trypsin) and the peptide amino acid sequence coverage (x-axis) of their experimental peptides. Four single-nucleotide polymorphism variants (rs10770, rs9898, rs2228243, and rs1042464) have been indicated by vertical black lines. Bottom panel: experimental peptides reported by the PeptideAtlas (accessed 16 May, 2017). The peptide amino acid sequence coverage (x-axis) is visualized in relation to experimental peptides for each experiment (y-axis). Highlighted in this example, rs9898 has only a few theoretical peptide sequences that can be observed with the bottom up method (because of sequence length) using this set of enzymes. Either one tryptic peptide with one missed cleavage NCPRHHFPR (identified once highlighted in the figure) or the chymotryptic peptide SCRNCPRH (based on low enzymatic specificity). For both panels, blue indicates the most abundant protein form and red is for any variation from the form.
