Abstract
Background
The emergence and spread of antimicrobial resistance (AMR) present a challenge to disease control in East Africa. Resistance to beta-lactams, which are by far the most used antibiotics worldwide and include the penicillins, cephalosporins, monobactams and carbapenems, is reducing options for effective control of both Gram-positive and Gram-negative bacteria. The World Health Organization, Food and Agricultural Organization and the World Organization for Animal Health have all advocated surveillance of AMR using an integrated One Health approach. Regional consortia also have strengthened collaboration to address the AMR problem through surveillance, training and research in a holistic and multisectoral approach. This review paper contains collective information on risk factors for transmission, clinical relevance and diversity of resistance genes relating to extended-spectrum beta-lactamase-producing (ESBL) and carbapenemase-producing Enterobacteriaceae, and Methicillin-resistant Staphylococcus aureus (MRSA) across the human, animal and environmental compartments in East Africa.
Main body
The review of the AMR literature (years 2001 to 2019) was performed using search engines such as PubMed, Scopus, Science Direct, Google and Web of Science. The search terms included ‘antimicrobial resistance and human-animal-environment’, ‘antimicrobial resistance, risk factors, genetic diversity, and human-animal-environment’ combined with respective countries of East Africa. In general, the risk factors identified were associated with the transmission of AMR. The marked genetic diversity due to multiple sequence types among drug-resistant bacteria and their replicon plasmid types sourced from the animal, human and environment were reported. The main ESBL, MRSA and carbapenem related genes/plasmids were the blaCTX-Ms (45.7%), SCCmec type III (27.3%) and IMP types (23.8%), respectively.
Conclusion
The high diversity of the AMR genes suggests there may be multiple sources of resistance bacteria, or the possible exchange of strains or a flow of genes amongst different strains due to transfer by mobile genetic elements. Therefore, there should be harmonized One Health guidelines for the use of antibiotics, as well as regulations governing their importation and sale. Moreover, the trend of ESBLs, MRSA and carbapenem resistant (CAR) carriage rates is dynamic and are on rise over time period, posing a public health concern in East Africa. Collaborative surveillance of AMR in partnership with regional and external institutions using an integrated One Health approach is required for expert knowledge and technology transfer to facilitate information sharing for informed decision-making.
Keywords: Genetic diversity, Risk factors, Antimicrobial resistance, Human-animal-environment, East Africa
Background
The emergence and the spread of antimicrobial resistance (AMR) presents a global challenge to diseases control. AMR can occur naturally following exposure to antimicrobial agents in the management of veterinary and human clinical cases [1]. The acquisition and spread of AMR can be attributed to ecological connectivity and presence in the community, of a previously acquired resistance gene, which can be denoted as a founder effect [2]. In resource-limited settings, the availability of antibiotics over the counter and without prescription mainly for self-treatment of suspected infections contributes to AMR. Of particular concern is the emergence of multi-drug resistant (MDR) bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) and extended spectrum beta-lactamase (ESBL)-producers. For instance, MDR invasive non-typhoidal Salmonella (iNTS) disease poses a major challenge to the clinical management of infections in resource-limited settings especially as alternative more effective antibiotics remain either unaffordable or simply unavailable for majority of patients [3].
In East Africa, resistance to commonly used antibiotics have been reported in humans [4–11], livestock [12, 13], wildlife [14] and environment [15, 16]. There is a worrying trend of increasing prevalence of AMR [17]. In recent years, the numbers of infections due to the ESBL-producing Enterobacteriaceae (ESBL-PE), MRSA, and carbapenem resistance (CAR) have increased, and they are now been recognized as “infections of concern” to East African and wider international populations. The carbapenem-based antibiotics, which are considered as “antibiotics of last resort” or “last-line agents”, have been compromised by the emergence of MDR Gram-negative bacteria [18]. In Sub-Saharan African countries, infection, and control measures against MRSA amplify the challenges in dealing with the AMR epidemic [19]. MRSA is primarily mediated by the mecA gene, located in staphylococcal cassette chromosome mec (SCCmec) that codes for a 78 kDa penicillin-binding protein (PBP2a), that has decreased affinity to methicillin and all beta-lactam antibiotics [20, 21]. Likewise, ESBL-PE possess a threat [22], associated with pediatric septicemia and urinary tract infections [23]. The largest ESBL group are the mutants blaTEM and blaSHV β-lactamases followed by blaCTX-M enzymes [24]; others include blaOXA-type β-lactamases, PER, VEB, GES.BES.TLA, SFO, IBC groups [24]. The co-emergence of ESBLs and carbapenemase-encoding plasmids facilitate selection for other resistance determinants for other antimicrobial classes, including aminoglycosides and fluoroquinolones, a key feature that fosters the spread of MDR in Enterobacteriaceae [25]. Further, a plasmid with ESBL determinants can also carry aminoglycosides and ciprofloaxcin resistance (CR) determinants. Such co-existence of resistance mutations on plasmids or ESBL related genes is not uncommon [26].
In East Africa, CR has been reported in Enterobacteriaceae and other gram-negative bacteria [9, 17, 27–32]. The high prevalence of CAR Enterobacteriaceae among Klebsiella pneumoniae and Escherichia coli present a challenge against treatment of the carbapenemase-producing Enterobacteriaceae infections [33]. Despite the fact that colistin is effective in the treatment of Gram-negative infections, it is toxic with a poor treatment outcome [34]. Resistance due to carbapenem antibiotics is mediated through the production of carbapenemases beta-lactamases enzymes such as veron integron metallo-beta-lactamases, imipenemase, K. pneumoniae carbapenemases, oxacillinase-48, and New Delhi Metallo-beta-lactamase_1, which are encoded by “blaVIM”, “blaIMP”, “blaKPC”, “blaOXA-48”, and “blaNDM” genes respectively [9].
In resource-limited settings, knowledge of the burden, distribution, and diversity of resistance genes in humans, animals and the environment is scarce [35]. Despite increasing research on AMR in such settings, few studies have adopted an integrated “One Health” (OH) approach to understand the transmission dynamics of AMR across humans, animals and the environment. Most AMR studies in resource-limited settings are human-focused [36], and constitute the majority of current knowledge [35]. Nevertheless, major policy reforms have been implemented in developing countries to generate a collective response to address AMR and antimicrobial use, set within a holistic and multisectoral One Health framework. In Tanzania, the SACIDS Foundation for One Health has focused on the burden of AMR in the southern African region, and has been providing support in the local and global fight against AMR. The SACIDS Foundation for One Health in partnership with the American Society for Microbiology implemented the Tanzania National Action Plan on AMR through the Fleming Fund Initiative to support AMR surveillance strategy. This strategy attempts to address the major gaps in AMR data collection and analysis, whilst strengthening antimicrobial stewardship. Furthermore, the Africa Centers for Disease Control and Prevention Framework for AMR in collaboration with public health institutions and leaders from human and animal health sectors have established a network for the surveillance of AMR. This network aims to measure, prevent, and mitigate the effects arising from drug resistance organisms.
Our paper presents and analyzes information on AMR from published articles in East Africa in order to provide an overview of risk factors for AMR transmission and genetic diversity of ESBL, MRSA and CAR resistance genes, as well as a consideration of the role of humans, animals and the environment in emergence and spread of AMR. Further, the review paper addresses the clinical relevance of ESBL, MRSA and CAR bacteria and initiatives which have been taken by regional consortia. These initiatives, in collaboration with external institutions, are addressing the AMR problem through an integrated One Health approach, and providing evidence-based information for decision making in the management of AMR clinical cases.
Materials and methods
The review of the literature was performed using search engines such as PubMed, Scopus, Science Direct, Google, Web of Science. Literature search was extended to involve repositories and sector ministries websites; however, no additional information was found. Search terms included ‘antimicrobial resistance and human-animal-environment, risk factors and predictors’, ‘antimicrobial resistance in human and animal populations’, prevalence and genotypic characterization of antimicrobial resistance’, methicillin-resistant Staphylococcus aureus’, ‘beta-lactamase genes and phenotypes’ Carbapenems resistance genes; ‘antimicrobial resistance genetic diversity and human-animal-environment’ combined with respective countries in East Africa including Burundi, Kenya, Rwanda, Tanzania and Uganda. The inclusion criteria included articles published between January 2001 and December 2018, which characterized AMR genes using genotypic methods in animals, humans and the environment. The exclusion criteria involved studies that did not utilize genotypic methods to identify resistance genes and sequence types of the AMR genes in the bacterial isolates.
Non-English language articles from French Speaking African countries such as Rwanda and Burundi were also excluded. The data were extracted using a template set of characteristics, including bacteria species, their origin (e.g. human, animal and environmental compartments), the proportion of the resistance genes, genotypic methods used for identification of AMR genes, sequence/clones/lineage types, plasmid replicon type and country of isolation. Further, descriptive statistics was performed to indicate trends of ESBLs, CARs and MRSA genes for the screened articles. The proportions of AMR genes from various pathogens in humans, animals and environment were correlated all-over the time-period for the screened studies using STATA version 14 (StataCorp LP, College Station, TX, USA), taking into consideration the time-period of recovered isolates. A p-value of < 0.05 considered statistically significant.
Results
The risk factors for emergence and transmission of AMR in animals, humans and the environment
The risk factors associated with the transmission of AMR genes varied within and between countries (Table 1), and can be broadly divided into five major types: (a) human practices, (b) demographic factors, (c) history of diseases and co-morbidities, (d) antibiotic use, and (e) hospitalization. We consider each in turn.
Table 1.
Risk factors for the emergence and transmission of AMR in East Africa
| Antimicrobial resistance study | Risk factors investigated | Significant risk factors | Hosts | Country | Reference |
|---|---|---|---|---|---|
| Nasal carriage of methicillin-resistant Staphylococcus aureus among healthy under-5 children | age, sex, education, visit the hospital, antibiotic use | – | human | Tanzania | [37] |
| Multiple ESBL-Producing E. coli Carrying Quinolone and Aminoglycoside Resistance Genes | sex, location, animal type, breed, antibiotic use | animal type, breed, antibiotic use | animals | Tanzania | [12] |
| Resistant E. coli in three culturally diverse ethnic groups | Increased number of water sources, adherence to antibiotic withdrawal periods, shared water resources, consumption of unboiled (raw) milk, | increased number of water sources, shared water, consumption of unboiled (raw) milk | Human, animals | Tanzania | [38] |
| Antibiotic Resistance in E. coli | Higher-income, antibiotics use | higher income | human | Tanzania | [39] |
| Nasal Carriage of Methicillin-Resistant Staphylococcus aureus | Duration in health care services, history of antibiotic use, history of chronic illness, duration in health care services, profession, age, location of health facilities, wards, | location of health facilities, duration in health care services | human | Tanzania | [40] |
| Extended-Spectrum-Beta-Lactamase-producing Enterobacteriaceae | Prior admission, prior medication, currently admitted in the surgical ward, patient inside the room, over 4 days of hospitalization, currently on the antibiotic, currently on Ciprofloxacin, currently on Ceftriaxone, HIV positive, wound infection | prior admission, currently on the antibiotic, wound infection, | human | Tanzania | [41] |
| Commonly used antimicrobial agents in bacterial pathogens isolated from urinary tract infections | Age, inpatient, hospitalization in the last 12 months, UTI in last 12 months, Urinary catheter, urinary catheter in last 12 months, use of other Antibiotics in the previous 6 months, Ciprofloxacin use in the previous 6 months, third-generation Cephalosporin use in the previous 6 months | Hospitalized (inpatient), third-generation Cephalosporin use in the previous 6 months, ciprofloxacin used in the previous 6 months | human | Rwanda | [42] |
| Antimicrobial resistance patterns of phenotype Extended Spectrum Beta-Lactamase producing bacterial isolates | Age, sex, department, sample type, ward/clinic, bacteria isolate, condition at discharge, the period of admission (days), | longer hospital stay, condition at discharge | human | Tanzania | [43] |
| Antimicrobial susceptibility profiles of E. coli and K. pneumoniae | Age, sex, health centre level, location of health sub-district, HSD, history of admission, history of medical procedures, surgery, antibiotic use, use of gentamicin, use of ciprofloxacin, use of septrin | age, level of health facility, location of health sub-district, district of residence, undergoing medical procedures, use of septrin | human | Uganda | [10] |
| Faecal carriage of ESBL-Producing Enterobacteriaceae | Sex, age, place of residence (district), parent level of education, children groups, hospitalized children, nutritional status, weight-for-age-Z-score, Length-for-age-Z-score, eight-for-length-Z-score, use of antibiotics, HIV | younger age, HIV infection and use of antibiotics | human | Tanzania | [44] |
| Predictors of blaCTX-M-15 in varieties of E. coli genotypes from humans | Age, number of children, sex, location, antibiotic use, admission history, | age, history of antibiotic use, history of admission in the past 1 year | human | Tanzania | [45] |
| Faecal carriage of CTX-M extended-spectrum beta-lactamase-producing Enterobacteriaceae | Source of income, source of food, local herbal use, street children type, primary education | local herbal use, street children type | human | Tanzania | [46] |
| Antimicrobial Resistance Profiles and Clonal Relatedness of Pseudomonas aeruginosa Strains | age, gender, residence, antimicrobial source, ease in accessing over the counter, prescription availability, dose completion | self-medication, non-completion of dosage | human | Kenya | [47] |
| Methicillin-resistant staphylococcus aureus (MRSA) colonization among Intensive Care Unit (ICU) patients and health care workers | Age, sex, education, occupation, smoking habit, history of sickness in past year, being sick for more 3 times, being diabetic, illicit drug use, | sex, history of sickness in past year, being sick for more 3 times, being diabetic, illicit drug use | human | Tanzania | [48] |
| Inappropriate usage of selected antimicrobials | Sex, age, breed, place/origin (rural, urban) | place/origin (rural/urban), age, breed | animals | Uganda | [49] |
| Extended-spectrum-beta lactamases producing Enterobacteriaceae | Age in days, sex, admission, body temperature, oxygen saturation, skin pustule, umbilical discharge, history of antibiotic-baby, maternal fever, maternal antibiotics, stool ESBL | positive ESBL-PE colonization of the mother, history of antibiotic use, | human | Tanzania | [50] |
Human practices
Transmission of AMR has been associated with practices involved in food production and animal husbandry [1]. Such practices include the use of antimicrobials in feeds as growth-promoters and to prevent infections [51–53] . For instance, Basulira et al. [49] reported a higher median concentration of antimicrobials such as beta-lactam antibiotics in adult carcass beef compared to young carcass beef samples. Such high levels of beta-lactam in feeds might be attributed by the indirect use of β-lactam antibiotics and tetracycline in feeds, drenches, drinking water and feed additives in the fattening systems [49]. In the Maasai community, consumption of unboiled (raw) milk was associated with increased odds of carriage of E. coli resistant to single and multiple antibiotics [38]. The relationship between milk and AMR was linked to consumption of antibiotic-resistant bacteria in contaminated milk [38]. Nevertheless, the Maasai and other communities are at a higher risk of diarrhea and other infections, from other sources. They tend to be greater consumers of certain types of antibiotics, leading to specific types of AMR [36]. In general, the spread of drug resistance is driven by multiple factors, which are linked to human life activity and travel, animals and the food trade, wild animals, migration, transportation, as well as water and wind [54].
Demographic factors
Demographic analysis of AMR is important to reduce any potential adverse health impact in a timely fashion to limit the spread of the resistant pathogens. In a human context, various studies have found an increased prevalence of beta-lactamase genes with age [37, 44, 45, 55]. In the community settings in Mwanza City, Tanzania, increasing age was reported as a predictor for the carriage of ESBL-PE [45]. Similarly, Muvunyi et al. [42] found an association between increased age with the prevalence of blaCTX-M Gram-negative bacteria. Likewise, an increased carriage of blaCTX-M Gram-negative bacteria in persons over 65-year-old due to repeated hospitalization has been reported [55]. Further, it was hypothesized that due to reduced immunity in adults over 65, they are more prone to infections caused by AMR bacterial strains [55]. In Uganda, the carriage of MDR isolates was significantly associated with age, particularly the age group between 15 and 44 years [10]. However, contrary to that, Tellevik and colleagues reported age equal to or below 12 months to be associated with ESBL carriage [44]. Furthermore, Tellevik and others reported that infants compared to older children were more likely to be colonized with ESBL positive bacteria, with the possibly that resistant strains were being transmitted from mothers to their babies [44].
History of co-morbidities
A history of malnourishment, diseases such as diabetes, HIV and other infections, were found to be associated with colonization of AMR bacteria [44, 48]. For instance, diabetic patients have reduced immunity, predisposing such patients to MRSA colonization [48]. Similarly, individuals with HIV are more prone to infections and hence are more likely to be hospitalized and consume antimicrobials [44]. Similarly, Cotton et al. [56] reported the association between HIV and ESBL carriage. The weak immunity in HIV patients increases the opportunity to acquire various infections [57]. Therefore, HIV positive pregnant women have been recommended to use anti-retroviral drugs to reduce mother-to-child transmission [44]. Despite the association between HIV and ESBL carriage, the HIV status has not been well documented as a risk factor for ESBL carriage [44]. Further, malnourished children with impaired immunity are more vulnerable to infections, and therefore more likely to be treated with antibiotics [44]. Godfrey and colleagues reported increased colonization of superficial skin infection with MRSA due to reduced skin immunity to fight against MRSA isolates [48].
History of antibiotics use
The previous use of antibiotics has been associated with the development of AMR [10, 12, 42, 44, 45, 50, 55]. The history of antibiotics use is the main driver in the development of AMR and hence selection pressure specific for the type of antibiotic and the bacterial species [55, 58–60]. In Tanzania, the history of antibiotic use and positive ESBL-PE colonization of mothers was associated with neonatal ESBL colonization in a tertiary hospital [50]. Moreover, a history of antibiotic use increased the risk of developing antibiotic resistance, where patients in Tanzania who were on treatment to at least one antibiotic had an increased risk of blaCTX-M Gram-negative bacteria compared to those who were not on any antibiotics [55]. In Rwanda, previous use of ciprofloxacin and other antibiotics in the previous 6 months was a risk factor contributing to CR [42]. Such findings also have been reported in other sub-Saharan African countries, particularly in Cameroon and Guinea-Bissau, where previous use of antibiotics was associated with ESBL carriage in hospital settings [61, 62]. However, this report is contrary to Moremi and colleagues who reported non-significant relationship between antibiotic use and ESBL-producing Enterobacteriaceae carriage among street children dwelling in Mwanza city [46]. Such a relationship was also reported by Fleece et al. [39], who found non-significant association between antibiotic use or episodes of diarrhea and antibiotic resistance in rural settings in Tanzania. In general, if the ecological and cultural conditions favour bacteria transmission, AMR bacteria could emerge irrespective of antimicrobial use practices in human health [38]. However, the heterogeneity between studies on the association between the previous use of antibiotics and AMR may be attributed to differences in study designs and the statistical methods used in data analysis.
Hospitalization
Prolonged stay in hospital and condition at discharge were significantly associated with ESBL producers [43]. The risk for AMR is increased especially in patients who had continued exposure to antibiotics during hospitalization [43]. For instance, Moremi and her colleagues reported that the ESBL-producing Enterobacteriaceae carriage was significantly higher on discharge than admission [63]. In other studies, being male, a history of sickness in the past year, and > 3 illnesses within a short time, were associated with MRSA colonization among patients [48]. Further, the level of available healthcare and facilities predisposed patients to AMR [10]. For instance, patients admitted at referral hospital such as Muhimbili National Hospital had higher ESBL carriage upon admission than those admitted to a District Hospital in Dar es Salaam [44]. This finding is also supported by Seni et al. [64] in which the resistance to third-generation cephalosporins in E. coli, Klebsiella spp., and other Enterobacteriaceae was higher in strains from a tertiary hospital compared to lower healthcare facilities. Similarly, at Bugando Medical Centre tertiary hospital in Mwanza, Tanzania, the MDR was higher in isolates from children than those from the district and regional hospitals [65]. In Moshi region, Tanzania, patients who came to the referral hospital at Kilimanjaro Christian Medical Centre were carrying more resistant bacteria than hospitalized patients [55]. This difference increased the risks to health workers, particularly nurses who had frequent patient contact, especially when compared to doctors [40]. Nevertheless, the association between MRSA carriage among healthcare workers and facilities might be attributed to differences in levels of commitment to control measures of infection between tertiary and regional hospitals [40]. For example, Moremi et al. [66], reported similarity of genotypes in the intensive care units due to frequent movement of healthcare workers or instruments, implying the need to revise cleaning and disinfection protocols, as well as having the necessary precautions to avoid hand and clothing contamination during clinical practices.
Genetic diversity, clinical relevance and regional surveillance of AMR in East Africa
Genetic diversity of AMR genes
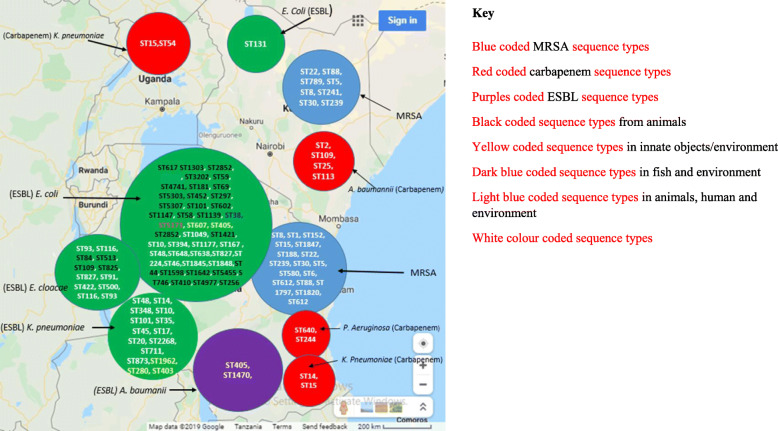
Most studies in East Africa have reported considerable genetic diversity and differences in sequence types of AMR genes particularly for ESBL (Table 2, Fig. 1), MRSA (Table 3) [71–75], and CAR (Table 4). These are attributed to evolutionary events, including mutations, selection and gene transfer, in the AMR genes in MRSA, ESBL and carbapenem isolates. In East Africa, ESBL resistance genes have been isolated from E. coli, Klebsiella spp., Proteus spp., Enterobacter spp., Pseudomonas aeruginosa, Acinetobacter spp., Klebsiella oxytoca, Proteus mirabilis, Enterobacter cloacae, Acinetobacter baumanii, Salmonellae spp. and E. cloacae complex [6–8, 12, 15, 22, 23, 44, 50]. Further, proportions and range of antimicrobial resistance genes obtained from human, animals and environment differ (Table 5).
Table 2.
The genetic diversity of extended–spectrum beta-lactamase (ESBL) genes
| Pathogen | Source | Genotypic tools | Antimicrobial resistance genes | Sequence types/clones | Plasmid Replicon type | Phylogroups | Country | Reference | Time period for collection of isolates |
|---|---|---|---|---|---|---|---|---|---|
| E. coli | chickens | PCR | blaTEM (100%), blaOXA-1 (75.0%), blaCMY-2 (62.5%), blaCTX-M-8 (50%), blaCTX-M − 9 (37.5%, blaCTX-M − 1,15 (12.5%), blaSHV (12.5%) | – | – | Tanzania | [67] | 2016 | |
| E. coli | pigs, cattle, sheep, goats, dogs, chicken | WGS, MLST | blaOXA-1 & blaTEM-1B (32%, 8/25), blaOXA-1 (2/25, 8%), blaTEM-1B 60% (15/25) | ST617, ST1303, ST2852, ST131, | IncFIA, IncFIB, IncFII, IncY,B/O/K/Z, IncX1, IncQ1, IncX3, IncX4, IncFIB(K), IncFIA | A, B1, B2, D | Tanzania | [12] | August and September 2014 |
| K. pneumoniae | human | PCR, WGS, AFLP | blaTEM-63 (4/9;44.4%), blaSHV-12 (0%), blaSHV-2a (0%), blaCTX-M-15 (5/9;55.6%), | – | – | Tanzania | [23] | August 2001 to August 2002, | |
| E. coli | human | PCR, WGS, AFLP | blaTEM-63 (4/25, 16%), blaSHV-2a (0/25, 0%), blaSHV-12 (0/25, 0%), blaCTX-M-15 (5/25, 20%) | – | – | Tanzania | [23] | August 2001 to August 2002, | |
| Salmonellae | human | PCR, WGS, AFLP | blaTEM-63 (3/17, 17.6%), blaSHV-2a (2/17, 11.8%), blaSHV-12 (2/17, 11.8%), blaCTX-M-15 (1/17, 5.9%) | – | Tanzania | [23] | August 2001 to August 2002, | ||
| K. pneumoniae | Human (blood, wounds, urine) | PCR, WGS, PFGE, MLST | blaCTX-M-15 (70/92; 76%), blaTEM-1, blaTEM-104 (18%), blaSHV-11 (3.2%), blaTEM-176 (2%) | ST48, ST14, ST348, ST10, | IncFII, IncND, IncFIA | – | Tanzania | [22] | Between April 2009 and March 2010 |
| E. coli | cattle | ERIC-PCR, WGS | blaTEM-1B, blaOXA-1, blaTEM-1A | ST1139, ST617, ST3202, ST59, ST4741, ST181, ST69, ST5303, ST452, ST297, ST5307, ST101, ST602, ST1147, ST58 | IncFIB (AP001918), ColRNAI, IncFIA, IncFII, IncFIC(FII),, InQ1, IncP, IncFII(pCoo), IncB/O/K/Z, Col156, IncFIB(pB171), IncFIA(HI1),, IncFII (pSE11),, IncX1, IncR, Incl1, Col(MG828), | A, B1, D | Tanzania | [13] | 2014 |
| E. coil | human | ERIC-PCR, WGS | blaTEM-1B, blaOXA-1, blaTEM-1A | IncFIB (AP001918), ColRNAI, IncFIA, IncFII, ColBS512, IncFII(pCoo), IncFII(29), IncFIB(K),lnO2,IncX4, Col(MP18),Col8282, | – | Tanzania | [13] | 2014 | |
| K. pneumonia | environment | PCR | blaCTX-M-1 | – | – | Tanzania | [15] | February 2014 | |
| E. coli | environment | PCR | blaCTX-M-1 (100%), blaCTX-M-9(5.9), other than blaCTM-1 & blaCTM-9 (17.6%) | – | – | Tanzania | [15] | February 2014 | |
| K. pneumoniae | Human (blood) | WGS | blaCTX-M-15, blaSHV-1, blaTEM-1B, blaSHV-11, blaSHV-33, blaSCO-1, blaOXA-1, blaSHV-28, blaSHV-83, blaSHV-27, | ST101, ST348, ST35, ST45, ST14, ST17, ST20, ST2268, ST711, ST873 | IncFIA, IncFIB, IncR, IncFII, IncHI1B, IncFR | – | Tanzania | [50] | Between July and December 2016 |
| E. cloacae | human | WGS | blaCTX-M-15, blaTEM-1B, blaOXA-1, blaACT, blaACT-7 | ST93, ST116, | IncHI2A, IncHI2, | – | Tanzania | [50] | Between July and December 2016 |
| A. baumanii | human | WGS | blaADC-25, blaNDM-1, blaOXA-69, blaOXA-58, blaCARB-8, | ST405, ST1470 | – | – | Tanzania | [50] | Between July and December 2016 |
| E. coli | human | RT PCR, WGS | blaCTX-M-15, blaCTX-M-14, blaCMY-2 | – | – | – | Tanzania | [44] | From August 2010 to July 2011 |
| K. pneumonia | human | RT PCR, WGS | blaCTX-M-15, | – | – | – | Tanzania | [44] | From August 2010 to July 2011 |
| K. oxytoca | human | RT PCR, WGS | blaCTX-M-15, | – | – | – | Tanzania | [44] | From August 2010 to July 2011 |
| E. cloacae complex | human | RT PCR, WGS | blaCTX-M-15, | – | – | – | Tanzania | [44] | From August 2010 to July 2011 |
| E. cloacae | inanimate surfaces and objects | PCR, DNA sequencing | – | ST84, ST513, ST109, ST825, ST827 | – | – | [66] | Between December 2014 and September 2015, | |
| Citrobacter spp | human | RT PCR, WGS | blaCTX-M-15, | – | – | – | Tanzania | [6] | between 1992 and 2010 |
| E. coli | human | PCR, WGS | blaTEM-1, blaSHV-1, blaCTX-M-14, blaCTX-M-15, blaCTX-M-9, blaCTX-M, blaCTX-M-3, blaCTX-M-1, blaSHV-5, blaSHV-12, blaTEM-52, blaTEM-125, blaTEM-50, blaTEM-78, blaTEM-109, TEM − 152, blaTEM-158, blaTEM-103, blaCMY-2, blaCMY-1, blaOXA-1 | – | – | Kenya | [68] | September and October 2009 | |
| E. coli | dogs, cats, | PCR, WGS, multiplex PCR | blaCTX-M-15 (dogs,47/216;22%), blaCTX-M-15(cats, 2/50;4%), blaOXA-1(dogs,47/216;22%), blaOXA-1 (cats, 2/50;4%), | ST131 | IncFIA, IncFIB, Incl1, IncFIA/FIB, | A, B1, B2 | Kenya | [68] | September and October 2009 |
| E. coli | human | PCR, multiplex PCR, WGS | blaCTX-M-154 (4/23;17%), blaOXA-1(4/23;17%) | ST131 | IncFIB | – | Kenya | [68] | September and October 2009 |
| E. coli | human | PCR | blaCTX-M, blaTEM | – | – | – | Kenya | [7] | March 2009 to February 2010 |
| K. pneumoniae | human | PCR | blaCTX-M, blaSHV, blaTEM | – | – | – | Kenya | [7] | March 2009 to February 2010 |
| E. coli | human | PCR, WGS | blaCTX-M-15, blaCMY-2 | – | – | – | Uganda | [41] | May 2010 to July 2011 |
| E. coli | human | PCR | blaCTX-M, blaSHV, blaTEM | – | – | – | Uganda | [41] | May 2010 to July 2011 |
| E. coli | human | PCR | blaCTX-M, blaSHV, blaTEM | – | – | Uganda | [41] | May 2010 to July 2011 | |
| E. coli | Fish | PCR | blaTEM (52%), blaSHV (36%), blaCTX 9.7%. | – | Tanzania | [47] | 2017 | ||
| E. coli | Fish | WGS | blaCTX-M-15 (9/11;81.8%), blaOXA-1 (6/11;54.5%), | ST-38, ST-5173 | IncI1, IncY, | E, B1 | Tanzania | [69] | between July and September 2015 |
| E. coli | Environment | WGS | blaCTX-M-15 (12/13;92.3%), blaOXA-1 (1/13;7.7%) | ST38, ST-2852, ST-1049, ST-1421, ST-131, ST-10, ST-394, ST-1177, ST-58, ST-167, ST-48, ST-5173 | IncY, IncI1, IncP, IncFII, IncFIA, IncFIB, IncQ1 | B1, A, B2, E, | Tanzania | [69] | between July and September 2015 |
| E. coli | inanimate surfaces and objects | PCR, DNA sequencing | blaCTX-M-15 | ST607, ST405 | – | – | Tanzania | [66] | December 2014 and September 2015, |
| K. pneumoniae | inanimate surfaces and objects | PCR, DNA sequencing | blaCTX-M-15 | ST1962, ST280, ST403 | – | – | Tanzania | [66] | December 2014 and September 2015, |
| E. cloacae | Fish | WGS | blaCTX-M-15, blaTEM-1B, blaACT-15, blaOXA-1, blaMIR-3, | ST91, ST422, ST500 | IncFII,IncFIB, IncFIB(K), IncFII, IncR | – | Tanzania | [69] | between July and September 2015 |
| K. pneumoniae | Fish | WGS | blaCTX-M-15, blaOXA-1, blaSHV-11, blaTEM-1B, | – | IncFII,IncFIB(K),IncHI1B, | – | Tanzania | [69] | between July and September 2015 |
| E. coli | Animals | WGS |
ST256, ST1303, ST1421, ST617, ST38, ST131, ST44, ST1598, ST1642, ST2852, ST5455, ST746, ST410, ST4977 |
IncFIA, IncFIB, IncFII, IncY,B/O/K/Z,IncX1, IncQ1, IncX3,IncIFIA, X4, IncFIB(K), | B1, A, D, B2, | Tanzania | [12] | between August/September 2014 | |
| E. coli | human | WGS | blaCTX-M-15, blaOXA-1, blaTEM-1B, | ST131, ST405, ST617, ST648 | IncFIA, IncFIB, IncFII, IncI2, IncI1, Col156, IncQ1, IncY, IncQ, Col (BS512) | – | Tanzania | [50] | Between July and December 2016 |
| A. baumanii | human | WGS | blaADC-25, blaOXA-69, blaNDM-1, blaOXA-58 | ST405, ST1470, | – | – | Tanzania | [50] | Between July and December 2016 |
| E. cloacae | human | WGS | blaCTX-M-15, blaTEM-1B, blaOXA-1, blaACT, blaACT-7 | ST116, ST93 | IncHI2A, IncHI2 | – | Tanzania | [50] | Between July and December 2016 |
| K. pneumoniae | human | WGS | blaCTX-M-15, blaSHV-1, blaTEM-1B, blaSHV-11, blaSHV-33, blaSCO-1, blaOXA-1, blaSHV-28, blaSHV-83, blaSHV-27, | ST101, ST348, ST35, ST45, ST48, ST14, ST17, ST20, ST2268, ST711, ST873 | ncFIA, IncFIB, IncR, IncFII, IncHI1B, | Tanzania | [50] | Between July and December 2016 | |
| E. coli | human | PCR, DNA sequencing | blaCTX-M-15, blaTEM-1 | ST131, ST405, ST638, ST38, ST827, ST224, ST648, ST46, ST1845, ST1848 | ncFIA, ncFIB, ncFII, ncFrepB, ncFIA- FIB | B2, D | Tanzania | [70] | 2011 |
| K. pneumoniae | human | WGS | blaSHV, blaLEN, blaOKP, blaTEM, blaCTX-M-15, blaOXA, blaSCO, blaDHA1, blaCARB, blaNDM-1 | ST15, ST54, | – | – | Kenya | [27] | 1994–2002 |
WGS Whole genome sequencing
*AFLP Amplified Fragment Length Polymorphism
*PFGE Pulse field gel electrophoresis
*MLST Multi-Locus sequence typing
*ERIC-PCR Enterobacterial Intragenic Consensus-Polymerase Chain Reaction fingerprinting
Fig. 1.
Geographical distribution of the sequence types from pathogens originated from various sources in the East Africa
Table 3.
Genetic diversity of Methicillin Resistance S. aureus (MRSA) in the East Africa region
| Pathogens | Source | Genotyping tools | Proportion of MRSA genes, n (%) |
Phylogroups/sequence types | Lineages/spa type | Country | Reference | Time period for collection of isolates |
|---|---|---|---|---|---|---|---|---|
| MRSA | human | WGS, MLST | mecA (10/30;33.3%) | 13 sequence types (ST-8, ST-1, ST-152, ST-15, ST-1847, ST-188, ST-22, ST-239, ST-30, ST-5, ST-580, ST-6 | Tanzania | [71] | August 2013 to August 2015 | |
| MRSA | human | Multiplex PCR | SCCmec I (1/69;1.4%), SCCmec II (52/69; 75.4%), SCCmec IV (2/69; 2.9%) | – | Kenya | [72] | 2005–2007 | |
| MRSA | human, environment | Multiplex PCR | SCCmec I (22/41;54%), SCCmec V (6/41;15%), SCCmec IV (3/41;7%) | – | Uganda | [73] | November, 2009 and February, 2010 | |
| MRSA | human | PCR, MLST | SCCmecA (24/24:100%) | ST88, ST 1797, ST1820 | t064, t104, t1855, t186, t667, t690, t7237, t7231, | Tanzania | [74] | Between January and December 2008 |
| MRSA | human | PCR, MLST | SCCmec type V (3/30;10%), SCCmec type IV (12/30;40%), SCCmec type III (13/30;43.3%), SCCmec type I (1/30;3.3%), SCCmec type II (1/30;3.3%) | ST22, ST88, ST789, ST5, ST8, ST241, ST30, | t037, t13149, t005, t022, t1339, t648, t345, t318, t293, t2029, t852, t689, t104, t1476, t13150, t091, t3202, t9622 | Kenya | [75] | Between January 2010 and July 2013 |
| MRSA | human | PCR, Multiplex PCR | SCCmec V (33.3%, 8/24), SCCmec IV (4.2% (1/24) SCCmec II (8.3% (2/24), SCCmec type 1 (16.7%, 4/24) | – | t645, t4353, t064, t355, t4609, t10277 | Uganda | [76] | September 2011 to April 2012 |
| MRSA | human | PCR, Multiplex PCR, WGS | SCCmec type I (56.4%), SCCmec type IV (17.9%) | – | – | Rwanda | [97] | |
| MRSA | human | WGS | SCCmec III (6/6; 100%) | ST239 | t037 | Kenya | [77] | Between the 11th July and 7th November 2011, |
| MRSA | cattle | PCR, Multiplex PCR, PFGE | SCCmec type V (21/23; 91.3%) | – | t7753, t1398, t2112, t3992, t127 | Uganda | [78] | July to August 2013 |
| MRSA | human | PCR, WGS | SCCmecA | ST612 | t690 | Tanzania | [66] | Between December 2014 and September 2015 |
| MRSA | human | PCR, WGS | SCCmec type IV (40.5%, 17/42), SCCmec type I (38.1%, 16/42), SCCmec types I, II and III (50%,21/42) | – |
t064 (19%, 8/42), t037 (12%, 5/42). t002, t037, t064, t4353 and t12939 |
Uganda | [16] | Between February and October 2011 |
Table 4.
Genetic diversity of Carbapenem-resistant genes in the East Africa region
| Pathogen | Source | Genotyping tools | Proportion of carbapenemase resistance genes | Phylogroups/sequence types | Lineages/cluster | Country | Reference | Time period for collection of isolates |
|---|---|---|---|---|---|---|---|---|
| P. aeruginosa | human, environment | PCR, WGS | blaIMP (36%, 9/25), blaVIM1 (32%, 8/25), blaSPM (16%, 4/25), blaNDM1 (4%, 1/25) | – | – | Uganda | [31] | Between February 2007 and September 2009 |
| A. baumannii | human, environment | PCR, WGS | blaOXA-24 (7%, 1/15), blaVIM-1 (13%, 2/15), blaOXA-58 (13%, 2/15), blaOXA-23 (60%, 9/15) | – | – | Uganda | [31] | Between February 2007 and September 2009 |
| P. aeruginosa | human | PCR, WGS, PFGE | blaVIM-2 (13.7%; 57/416), blaVIM-1 (13.7%; 57/416) | – | MBLA, MBLAR, MBLB, | Kenya | [32] | 2006 and 2007 |
| P. aeruginosa | human | PCR, WGS, PFGE | blaIMP (12/49;24.5%), blaVIM (9/28; 32.1%), blaOXA_48 (2/11;18.2%), blaKPC (1/8; 12.5%), blaNDM (1/8; 12.5%) | – | – | Tanzania | [9] | Between 2007 and 2012 |
| P. aeruginosa | human | PCR, WGS | blaVIM-2 | ST640, ST244 | Tanzania | [41] | May 2010 to July 2011 | |
| K. pneumoniae | human | PCR, WGS, PFGE | blaIMP types (49/227; 21.6%), blaVIM types (28/227; 12.3%), blaOXA_48 11/227 (4.9%), blaKPC (8/227; 3.5%), blaNDM (7/227;3.1%), blaNDM-1, blaIMP, blaNDM-1, | ST14, ST15 | – | Tanzania, | [9] | Between 2007 and 2012 |
| K. pneumoniae | human | PCR, WGS, PFGE | blaNDM-1 (4/35; 11.4%), blaVIM (16/35;45.7%), blaIMP (5/35;8.16%), blaKPC (3/35;8.6%), blaOXA-48 (7/35;20%), | – | – | Uganda | [17] | between January, 2013 and March, 2014 |
| K. pneumoniae | blaVIM (17/43;39.5%), blaOXA-48 (5/13;38.5%) | – | Uganda | [30] | January, 2013 and March, 2014 | |||
| E. coli | human | PCR | blaIMP (19/32;59.4%), blaNDM (0/32;0%), blaVIM (4/32;12.5%), blaOXA_48 (3/32;9.4%), blaKPC (4/32; 12.5%), blaNDM (2/32;6.3%) | – | – | Tanzania, | [9] | Between 2007 and 2012 |
| E. coli | human | PCR | blaNDM-1 (0/19; 0%), blaVIM (1/19;5.3%), blaIMP (6/19;31.6%), blaKPC (4/19;21.1%), blaOXA-48 (8/19;42.1%) | – | – | Uganda | [17] | between January, 2013 and March, 2014 |
| E. coli | human | PCR | blaVIM (20/43;46.5%), blaOXA-48 (6/13;46.2%) | – | – | Uganda | [30] | January, 2013 and March, 2014 |
| A. baumannii | human | PCR, PFGE, WGS | blaIMP types (3/3;100%),blaVIM (0/3; 0%)blaOXA_48 (0/3; 0%), blaKPC (0/3;0%), blaNDM (0/3;0%) | – | – | Tanzania, | [9] | Between 2007 and 2012 |
| A. baumannii | human | PCR, PFGE, MLST | ISAba1-blaOXA-23, blaOXA-51-like, blaADC, blaNDM-1, ADC-57. | ST2, ST109, ST25, ST113 | European clone II (ECII) | Kenya | [28] | January 2009 to August 2010 |
| Salmonella spp | human | PCR | blaIMP types (1/2;50%),blaVIM(1/50; 50%) | – | – | Tanzania | [9] | Between 2007 and 2012 |
| K. oxytoca | human | PCR | blaIMP types (3/5;60%),blaOXA481/5;20%),blaNDM1/5;20%) | – | – | Tanzania, | [9], | Between 2007 and 2012 |
| K. oxytoca | human | PCR | blaNDM-1 (1/1; 100%), blaVIM (0/1;0%), blaIMP (0/1;0%),blaKPC (0/1;0%), blaOXA-48 (1/1;100%) | – | – | Uganda | [17] | between January, 2013 and March, 2014 |
| C. freundii | human | PCR | blaIMP types (2/4;50%), blaVIM (1/4;25%), blaOXA_48 (1/4;25%), | – | – | Tanzania | [9] | Between 2007 and 2012 |
| C. freundii | human | PCR | blaNDM-1 (1/1; 12.5%), blaVIM (0/1;0%), blaIMP (0/8;0%), blaKPC (0/1;0%), blaOXA-48 (0/1;0%), | – | – | Uganda | [17] | between January, 2013 and March, 2014 |
| P. aeruginosa | human | PCR | blaIMP types (12/25;48%), blaVIM (9/25;36%), blaOXA_48 (2/25;8%),blaKPC (1/25;4%),blaNDM (1/25; 4%) | – | – | Tanzania | [9] | Between 2007 and 2012 |
| Enterobacter spp. | human | PCR | blaNDM-1 (1/5; 20%), blaVIM (1/5;20%), blaIMP (0/1;0%), blaKPC (1/5;0%), blaOXA-48 (2/5;40%) | – | – | Uganda | [17] | between January, 2013 and March, 2014 |
| Pantoea agglomerans | human | PCR | blaNDM-1 (0/1; 0%), blaVIM (0/1;0%), blaIMP (1/1;100%), blaKPC (0/1;0%), blaOXA-48 (0/1;0%) | – | – | Uganda | [17] | between January, 2013 and March, 2014 |
| Proteus mirabilis | human | PCR | blaNDM-1 (0/2; 0%), blaVIM (0/2;0%), blaIMP (0/2;0%), blaKPC (1/2;50%), blaOXA-48 (1/2;100%) | – | – | Uganda | [17] | between January, 2013 and March, 2014 |
| Proteus mirabilis | human | Multiplex PCR | blaVIM1/43;2.3%), blaOXA-48 (1/13;7.7), | – | – | Uganda | [30] | September 2013 to June 2014, |
| Serratia marcescens | human | PCR | blaNDM-1 (0/3; 0%), blaVIM (2/3;66.7%), blaIMP (0/3;0%), blaKPC (1/3;33.3%), blaOXA-48 (0/3;0%) | – | – | Uganda | [17] | between January, 2013 and March, 2014 |
| Morganella morganii | human | Multiplex PCR | blaVIM1/43; %2.3), blaOXA-48 (0/13;0%), | – | – | Uganda | [30] | September 2013 to June 2014, |
| Enterobacter sakazaki | human | Multiplex PCR | blaVIM 0/13; 0%),blaOXA-48 (1/13; %7.7), | – | – | Uganda | [30] | September 2013 to June 2014, |
| Stenotrophomonas spp | human | Multiplex PCR | blaVIM (1/43;2.3%),blaOXA-48 (0/23;0%) | – | – | Uganda | [30] | September 2013 to June 2014, |
Table 5.
Proportions and range of antimicrobial resistance genes obtained from various pathogens in humans, animals and environment
| Type of antimicrobial resistance genes | Range of ESBL genes in different studies (%) | Mean (n/N;%) |
|---|---|---|
| Methicillin resistanceS. aureusgenes | ||
| SSCmec I | 0–56.4 | 21.24 |
| SSCmec II | 0–8.3 | 1.45 |
| SSCmec III | 0–100 | 27.34 |
| SSCmec IV | 0–40.5 | 14.06 |
| SSCmec V | 0–91.3 | 18.70 |
| SSCmec I,II,III | 0–50 | 6.25 |
| SSCmecA | 0–100 | 16.66 |
| Extended spectrum beta-lactamase genes | ||
| TEM | 0–100 | 26.7 |
| OXA | 0–75 | 15 |
| CMY | 0–62.5 | 4.81 |
| CTX-M | 0–100 | 45.68 |
| SHV | 0–36 | 7.72 |
| OXA&TEM | 0–32 | 2.46 |
| Carbapenames genes | ||
| IMP | 0–100 | 23.75 |
| VIM | 0–66.7 | 18.84 |
| SPM | 0–16 | 0.64 |
| NDMI | 0–100 | 7.59 |
| OXA | 0–100 | 17.91 |
| KPC | 0–33.30 | 5.82 |
The genetic diversity of ESBL genes including blaTEM, blaOXA-1, blaCMY-2, blaCTX, blaSHV, blaSCO, blaACT, blaADC, blaCARB, and blaNDM have been collected from bacterial species sourced from animals, humans and the environment (Table 2). Such diversity of ESBL producing bacteria represents a huge challenge for local hospital infection control teams [22]. The ESBL, particularly blaTEM-, blaSHV-, and blaCTX M-enzyme, exhibit a high degree of diversity with high levels of MDR [31, 44, 79, 80]. In East Africa, the blaCTX-Ms have been reported to be the dominant enzymes among the ESBL genes [6–8, 15, 44, 69, 81]. However, this finding is contrary to reports by Armah [67] in Morogoro region, Tanzania in which the blaTEM enzymes followed by blaOXA-and blaCMY-2 were isolated in higher proportion in poultry. The ESBL are encoded by plasmids and mobile genetic elements and transferred to other bacteria [82]. The genetic diversity of plasmid replicons types in humans, animals and environmental compartments has been reported in E. coli, K. pneumoniae and E. cloacae ESBL carrying isolates (Table 2). The IncF plasmids have been found in E. coli producing ESBL genes collected from animals, humans and the environment [12, 13, 68, 69]. Further, the IncF plasmid types such as IncFIA, IncFIB, IncFII, IncFIB (K) have been reported in E. coli producing ESBLs in domestic and companion animals [12], humans [68], thereby indicating commonality of IncF plasmids types circulating in humans and animals. Such associations have also been reported in K. pneumoniae and E. cloacae ESBL carrying isolates originating from humans [22, 50, 69] and fish [69]. The IncF types such as IncFIB (AP001918), IncFIA, IncFII, IncFII (pCoo c) were shared between animals and humans [13], indicating the possibility of cross-species transmission of plasmid replicon type among ESBL-PE genes carrying isolates.
In East Africa, the distribution and the predominance of the MRSA genes (Table 3) varies between countries. In Uganda, the SCCmec type 1 and SCCmec type V were the most common and accounted for between “33.3 - 91.3%” prevalence [73, 76, 78]. In Kenya, the SCCmec type II MRSA and a pvl strain of MRSA were significant pathogens in patients with soft tissue infections presenting to hospitals [72]. Further, a cross sectional study in Kenya, reported predominance of SCCmec type V followed by SCCmecII genes in healthy students residing within the university residence halls [83]. Several sequence types (STs) of MRSA have been reported in East Africa [71, 74, 75, 77]. For instance, the ST88 and ST1797 have been reported to be the predominant sequence types causing wound infection and abscesses in clinical isolates in a tertiary hospital in Tanzania [74]. In Kenya, ST241 was the predominant clonal complex in various healthcare institutions in Nairobi [75]. Further, ST239 isolates were associated with hospital-acquisition in Kenya [77]. The MRSA carriage isolates have been confined to patients with burns due to a prolonged stay in the hospital [77]. MRSA isolates were recovered from wound swabs, pus, and nasal swabs in Lake Victoria, Tanzania, and found the spa types t690 and t7231, ST88 and ST1797 were dominant among clinical cases [74]. The two MRSA isolates from wounds were all typed as ST612 [11]. In Kenya, all the MRSA isolates collected from inpatients in the medical, surgical and gynecological wards of Thika Hospital were of spa type t037, linked to internal sources within the hospital [77].
A wide diversity of CAR genes including blaIMP, blaVIM-1 blaSPM-l, blaNDM-1, blaOXA-23 blaOXA-24, blaOXA-58, blaKPC and novel ADC, blaADC-57 have been identified in East Africa [28, 33] (Table 4), mostly isolated from clinical samples in hospital settings. However, there is no information available on occurrence of CP bacteria in livestock and their environment in the East Africa [33]. The genetic determinants of CP have been reported in P. aeruginosa, A. baumannii, K. pneumoniae, Salmonella spp., K. oxytoca, Enterobacter spp., Pantoea agglomerans, P. mirabilis, Serratia marcescens, Pantoea agglomerans, Morganella morganii, Enterobacter sakazaki and Stenotrophomonas spp. (Table 4). The prevalence of carbapenemase genes isolated from different bacterial isolates varies. For instance, Mushi et al. [9] reported variation in the prevalence of CP genes, in which the overall frequency of drug resistance loci in E. coli was 14% (32 isolates) followed by K. pneumoniae 10.57% (24 isolates), P. aeruginosa 10.13%, K. oxytoca 1.76%, A. baumannii 1.3%, Citrobacter freundii 0.88%, S. marcescens 0.88%, and Salmonella spp. 0.44%). In South Western Uganda only blaVIM and blaOXA-48 genes were detected among the carbapenemase-producing Enterobacteriaceae of clinical origin [30]. However, most studies have reported low prevalence of the blaNDM-types. For instance, the blaNDM-1 gene was found to be uncommon (2.6%) among the bacterial isolates [17], signifying the low prevalence of the New Delhi Metallo-β-lactamase 1 (NDM-1) types in the East Africa. The blaNDM-1 genes have been reported in Kenya [28], Uganda [17, 31] and Tanzania [9, 31]. The blaNDM-1 genes were for the first time isolated in India and then reported in Europe and have been identified in extensively drug-resistant A. baumannii [28], raising a concern in the transmission dynamics of these genes in East Africa. Poirel and colleagues reported identical PGFE patterns of K. pneumoniae similar to strain 05–506 identified in Sweden. Strain 05–506 was associated with intercontinental transmission of the blaNDM-1, by the Indian population in Kenya, and air traffic between Europe and East Africa [80]. The blaNDM-1 gene has been found co-existing with other CP genes, thereby explaining why these isolates are MDR [17]. In Kenya, the blaNDM-1 carbapenem-resistant NDM-1-positive K. pneumoniae isolates were clonally related and expressed other ß-lactamases genes including the blaCTX-M-15, blaOXA-1, blaOXA-9, blaCMY-6 and aminoglycoside resistance methylase RmtC genes [80]. Further, all isolates that carried the blaNDM-1 carbapenemase gene were clonally related and expressed many other resistance determinants, including β-lactamases blaCTX-M-15, blaOXA-1, blaOXA-9, blaCMY-6, and aminoglycoside resistance methylase RmtC [80]. In Tanzania, Mushi and colleagues found solitary and heterogenous MDR gram-negative bacteria having at least two carbapenemase genes [9].
Trends in the carriage of antimicrobial resistance genes of the recovered isolates overtime period
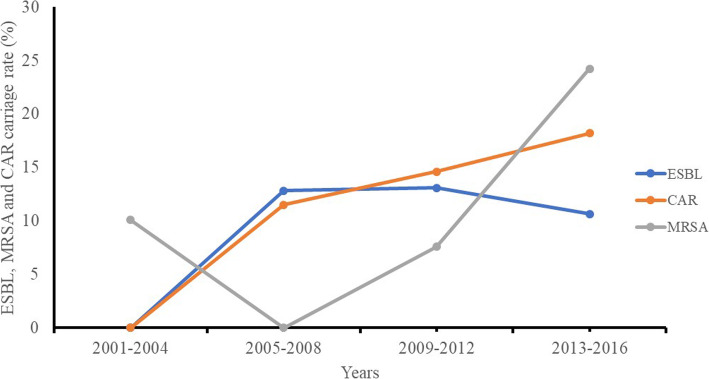
Figure 2 represents the ESBL, MRSA, and CAR carriage rates over time, as established by a linear regression model using the values reported in screened studies conducted from 2001 to 2018 in East Africa. Over this period, the differences in ESBLs, MRSA, and carbapenem carriage rate in screened studies in East Africa were insignificant (p-value = > 0.05). The data obtained with period on carriage rate between ESBL and CAR [p value = 0.1; 95% CI = − 0.5 to 1.9%], ESBL and MRSA (p value = 0.9; 95% CI = 1.9 to 1.8%] and CAR and MRSA; [p value = 0.6; 95% CI = − 1.9 to 2.5%] were insignificant. However, AMR gene carriage in recovered isolates is on the rise and increased from 2001 to 2016, 2005 to 2016, and 2001 to 2012 for CAR, MRSA, and ESBLs respectively. The highest AMR genes carriage rates for CAR (18.2%), ESBL (13.1%), and MRSA (24.25%) were reported in studies conducted from 2013 to 2016, 2009–2012, and 2013–2016 respectively. The rise of ESBL, CAR, and MRSA carriage rates in East Africa pose a threat to public health particularly in the geographical areas where the rates are very high. A limitation of this analysis is the fact that these data were collected from unlinked cross-sectional studies conducted in different geographical areas in East Africa; consequently, some important information may have been missed.
Fig. 2.
Trends in Extended Spectrum beta-lactamase, Methicillin resistance Staphylococcus aureus and carbapenem genes in recovered isolates in East Africa
Clinical relevance and outcome of the AMR genes in East Africa
The genetic diversity at antibiotic resistance loci could determine a population’s capacity to cope with future challenges of the antimicrobial drugs used for infection control [84]. The genetic diversities of AMR genes particularly ESBLs, MRSA, and CAR among clinical isolates in East Africa pose a public health threat. In East Africa, the ESBL, particularly TEM-, SHV-, CTX M-enzyme, exhibit a high degree of diversity [79, 80], associated with high levels of MDR [44]. The ESBLs are encoded by plasmids which also carries genes encoding for other drugs, limiting the antibiotic options in the treatment of ESBL producing organisms [85]. Carbapenems have been considered as the drug of choice for infection associated with ESBL-producing organisms. However, challenges remain as resistance to carbapenems has recently been reported, and there have been treatment failures [85]. The ESBL-producing organisms may appear susceptible to some extended-spectrum cephalosporins [85]. However, alternative treatment options to cephalosporins are limited in local settings. In Uganda, a high prevalence of carbapenem and CAR encoding genes among third-generation cephalosporins resistant Enterobacteriaceae have been reported in clinical isolates obtained from patients referred to Mulago hospital. Furthermore, high resistance rates in third-generation cephalosporin cefotaxime among urinary tract infection isolates have been recovered from pregnant women at Muhimbili National Hospital in Tanzania [86].
The high MRSA carriage rates among patients and health workers in the East Africa is a concern given poor infection prevention and control measures in our settings [87]. In Kenya, the trend in antibiotic susceptibility to MRSA declined between years 2014 to 2016, just like the methicillin susceptible counterparts [87]. In Kenya, MRSA is susceptible to linezolid, tigecycline, teicoplanin and vancomycin [87], quinupristin/dalfopristin, nitrofurantoin, and ampicillin/sulbactam [88]. Further, high resistance to commonly used antibiotics such as gentamycin, erythromycin, levofloxacin and tetracycline has also been reported in Kenya [88]. In Tanzania, high resistance rates among MRSA isolates to kanamycin, gentamicin, ciprofloxacin and trimethoprim-sulphamethoxazole have been reported [40]. Low resistance rates against MRSA isolates have been reported to mupiroaazcin and none toward linezolid, indicating that these two drugs can be used as a treatment options for MRSA infections in our settings [40]. In Uganda, the coexistence of hospital and community-associated MRSA have been fueled by pre-exposure to ampicillin and co-trimoxazole/or health care facilities in children [16]. Such coexistence presents a challenge for management of MRSA infection in outpatients.
Strategies and future for regional surveillance of AMR in the East Africa
The World Health Organization, the Food and Agriculture Organization of the United Nations and the World Organization for Animal Health has advocated for a holistic and multisectoral approach to address the AMR problem. This has been strengthened by the collaboration of regional consortia involving local and regional academic and research institutions in Africa, and a joint dedication of efforts to fight against infectious diseases in animals, humans and their environments. Regional consortia such as the SACIDS Foundation for One Health in collaboration with the London School of Hygiene and Tropical Medicine, London International Development Centre, Royal Veterinary College, Chatham House of the Royal Institute of International Affairs in the United Kingdom and the American Society of Microbiology of the United States, have partnered to address AMR problem through research and training. Strategically, surveillance is being supported by phenotypic and genomic data, to understand the flow of resistomes across human, animal and environmental compartments. For instance, research programs focused on genomic epidemiology of ESBLs producing E. coli in humans, animals and their environments are conducted by postgraduate students of the SACIDS Foundation for One Health at the Sokoine University of Agriculture and Muhimbili University of Health and Allied Sciences in Tanzania. This work is attempting to understand the transmission dynamics of AMR genes and their virulence [89], and will provide a model for the surveillance of AMR in Africa, including an exemplar of implementing global recommendations and local solutions. More generally, surveillance will be performed in line with the national AMR action plans to establish a nation-wide surveillance system for AMR, establish and build capacity for a national reference laboratory and designated laboratories for AMR surveillance using an integrated One Health approach. SACIDS Foundation for OH in collaboration with the American Society for Microbiology through the Fleming Fund initiative will implement the Tanzania National Action Plan for AMR launched in 2017. Further, through funding from the Medical Research Council in the United Kingdom, policy gaps, behaviour, socio-cultural and economic determinants for AMR in communities will be identified to provide evidence-based policy by policy makers to solve the AMR problem in Tanzania.
The implementation of effective and sustainable AMR surveillance programmes in Africa is hampered by a lack of infrastructure and other resources required to perform optimal surveillance [90]. Most laboratory facilities in resource-limited countries produce quality routine culture and susceptibility testing, which is both cost-effective and provides high-quality surveillance data. However, developing countries should consider incorporation of the whole genome sequencing in the diagnosis framework of drug resistance isolates. The MDR isolates present a challenge for management of clinical cases in resource-limited countries. Resistance to colistin, which is a last resort treatment for life-threatening infections caused by Enterobacteriaceae, including MDR pathogens has been reported in several countries and regions. Other infections such as MDR tuberculosis (MDR-TB) pose a threat to public health in the Sub Saharan African countries adding to the current long-standing pandemics of HIV in the region. Studies have found high diversity of mutations among MDR-TB patients, suggesting established and ongoing transmission of MDR-TB strains or multiple sources of infections [91]. However, it is unclear whether the role played by drug resistance genetic mutations in combination or alone could predict the disease manifestations or progression. Therefore, clinical studies have been proposed to evaluate the incorporation of whole genome sequencing in the diagnostic framework of drug resistance tuberculosis in these countries. It has been thought that the incorporation of whole genome sequencing as a diagnosis tool for MDR-TB in resource limited settings will improve the diagnosis and treatment of TB while providing stringent strain discrimination. Such incorporation would improve the detection of drug resistance mutations in TB patients, and to inform clinical and treatment decision making [92]. The information obtained from such clinical studies could support policy change in the adoption of molecular TB diagnostic algorithms in resource-limited countries.
Conclusions
The high genotypic diversity of the AMR genes among the bacterial isolates might suggest possible exchange of strains or a flow of genes among different strains due to transfer by mobile genetic elements or multiple sources of resistance bacteria. Such levels of diversity prompt the calling for immediate interventions, including guidelines concerning antibiotics use and regulations governing their importation and sale. However, antibiotic use and regulation is likely to be a very complex system because other factors such as cultural and ecological conditions favour transmission of bacteria. There is a high likelihood that people will have AMR bacteria irrespective of how antibiotics are used. Therefore, control strategies against AMR need to be tailored, beyond antibiotic use and availability, to local practices that affect bacterial transmission. Moreover, the trend of ESBL, MRSA and CAR carriage rates is dynamic and are on rise over time period, posing a public health concern in East Africa. In addition to that, phenotypic and genotypic drivers of AMR should be investigated within an integrated One Health framework. Such investigations will inform transmission dynamics of resistomes across compartments, and facilitate information sharing for informed decision making; ultimately, reducing the spread of AMR genes in bacteria. Further, the application of advanced techniques such as whole genome sequencing for detection of AMR could cut a bridge between clinical research and care, so that case management and treatment decisions can be informed and personalised.
Acknowledgements
The authors would like to thank the SACIDS Foundation for One Health and partner institutions in Tanzania and the United Kingdom for their support during the preparation of the manuscript.
Authors’ contributions
BZK, GM, SM, HC, TGC, SC, LM, MMM and MIM searched the literature and commented on the manuscript; BZK drafted the first manuscript. All authors read and approved the manuscript for submission.
Funding
This study received funding from the SACIDS Africa Centre of Excellence for Infectious Disease (SACIDS-ACE) through the Government of the United Republic of Tanzania and the World Bank (WB ACE II Grant P151847) to undertake a postdoctoral research fellowship at Muhimbili University of Health and Allied Sciences. TGC is funded by the Medical Research Council UK (Grant no. MR/M01360X/1, MR/N010469/1, MR/R025576/1, and MR/R020973/1) and the BBSRC (Grant no. BB/R013063/1). SC is funded by Medical Research Council UK grants (MR/M01360X/1, MR/R025576/1, and MR/R020973/1) and the BBSRC (Grant no. BB/R013063/1).
Availability of data and materials
Data and materials were available from peer reviewed articles published between January 2001 and December 2018 in East Africa.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Allcock S, Young EH, Holmes M, Gurdasani D, Dougan G, Sandhu MS, Solomon L, Török ME. Antimicrobial resistance in human populations: challenges and opportunities. Glob Health Epidemiol Genom. 2017;2:e4. doi: 10.1017/gheg.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martínez JL, Baquero F. Emergence and spread of antibiotic resistance: setting a parameter space. Ups J Med Sci. 2014;119(2):68–77. doi: 10.3109/03009734.2014.901444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kariuki S, Gordon MA, Feasey N, Parry CM. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine. 2005;33:C21–C29. doi: 10.1016/j.vaccine.2015.03.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mshana SE, Kamugisha E, Mirambo M, Chakraborty T, Lyamuya EF. Prevalence of multiresistant gram-negative organisms in a tertiary hospital in Mwanza, Tanzania. BMC Res Notes. 2009;2:49. doi: 10.1186/1756-0500-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kayange N, Kamugisha E, Mwizamholya DL, Jeremiah S, Mshana SE. Predictors of positive blood culture and deaths among neonates with suspected neonatal sepsis in a tertiary hospital, Mwanza- Tanzania. BMC Pediatr. 2010;10:39. doi: 10.1186/1471-2431-1110-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiiru J, Kariuki S, Goddeeris BM, Butaye P. Analysis of beta-lactamase phenotypes and carriage of selected beta-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC Microbiol. 2012;12:155. doi: 10.1186/1471-2180-1112-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maina D, Revathi G, Kariuki S, Ozwara H. Genotypes and cephalosporin susceptibility in extended-spectrum beta-lactamase producing enterobacteriaceae in the community. J Infect Dev Ctries. 2012;6(6):470–477. doi: 10.3855/jidc.1456. [DOI] [PubMed] [Google Scholar]
- 8.Moses A, Bwanga F, Boum Y, Bazira J. Prevalence and genotypic characterization of extended-Spectrum Beta-lactamases produced by gram negative bacilli at a tertiary Care Hospital in Rural South Western Uganda. Br Microbiol Res J. 2014;4(12):1541–1550. doi: 10.9734/BMRJ/2014/9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mushi MF, Mshana SE, Imirzalioglu C, Bwanga F. Carbapenemase genes among multidrug resistant gram-negative clinical isolates from a tertiary Hospital in Mwanza, tanzania. BioMed Res Int. 2014;2014:303104. doi: 10.1155/2014/303104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Najjuka CF, Kateete DP, Kajumbula HM, Joloba ML, Essac SY. Antimicrobial susceptibility profiles of Escherichia coli and Klebsiella pneumoniae isolated from outpatients in urban and rural districts of Uganda. BMC Res Notes. 2016;9:235. doi: 10.1186/s13104-016-2049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moremi N, Claus H, Vogel U, Mshana SE. The role of patients and healthcare workers Staphylococcus aureus nasal colonization in occurrence of surgical site infection among patients admitted in two centers in Tanzania. Antimicrob Resist Infect Control. 2019;8:102. doi: 10.1186/s13756-019-0554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seni J, Falgenhauer L, Simeo N, Mirambo MM, Imirzalioglu C, Matee M, Rweyemamu M, Chakraborty T, Mshana SE. Multiple ESBL-producing Escherichia coli sequence types carrying quinolones and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Front Microbiol. 2016;7:142. doi: 10.3389/fmicb.2016.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madoshi BP, Kudirkiene E, Mtambo MMA, Muhairwa AP, Lupindu AM, Olsen JE. Characterization of Commensal Escherichia coli Isolated from Apparently Healthy Cattle and Their Attendants in Tanzania. PLoS One. 2016;11(12):e0168160. doi: 10.1371/journal.pone.0168160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katakweba AA, Moller KS, Muumba J, Muhairwa AP, Damborg P, Rosenkrantz JT, Minga UM, Mtambo MMA, Olsen JE. Antimicrobial resistance in faecal sampls from buffalo, wildebeest and zebra grazing together with and without cattle in Tanzania. J Appl Microbiol. 2015;118(4):966–975. doi: 10.1111/jam.12738. [DOI] [PubMed] [Google Scholar]
- 15.Erb S, D’Mello-Guyett L, Malebo HM, Njee RM, Matwewe F, Ensink J, Hinic V, Widmer A, Frei R. High prevalence of ESBL-Producing E. coli in private and shared latrines in an informal urban settlement in Dar es Salaam, Tanzania. Antimicrob Resist Infect Control. 2018;7(1):1. doi: 10.1186/s13756-017-0292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kateete DP, Bwanga F, Seni J, Mayanja R, Kigozi E, Mujuni B, Ashaba FK, Baluku H, Najjuka CF, Källander K, et al. CA-MRSA and HA-MRSA coexist in community and hospital settings in Uganda. Antimicrob Resist Infect Control. 2019;8:94. doi: 10.1186/s13756-019-0551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okoche D, Asiimwe BB, Katabazi FA, Kato L, Najjuka CF. Prevalence and characterization of Carbapenem-resistant Enterobacteriaceae isolated from Mulago National Referral Hospital, Uganda. PLoS One. 2015;10(8):e0135745. doi: 10.1371/journal.pone.0135745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pop-Vicas A, Opal MS. The clinical impact of multidrug-resistance gram-negative bacilli in the management of septic shock. Virulence. 2014;5(1):206–212. doi: 10.4161/viru.26210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falagas ME, Karageorgopoulos DE, Leptidis J, Korbila IP. MRSA in Africa: filling the global map of antimicrobial resistance. PLoS One. 2013;8(7):e68024. doi: 10.1371/journal.pone.0068024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stryjewski ME, Ralph Corey G. Methicillin-resistant Staphylococcus aureus: an evolving pathogen. Clin Infect Dis. 2014;58(1):S10–S19. doi: 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- 21.Abdulgader SM, Shittu AO, Nicol MP, Kaba M. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol. 2015;6:348. doi: 10.3389/fmicb.2015.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mshana SE, Hain T, Domann E, Lyamuya EF, Chakraborty T. C I: Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis. 2013;13:466. doi: 10.1186/1471-2334-1113-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blomberg B, Jureen R, Manji KP, Tamim BS, Mwakagile DSM, Urassa WK, Fataki M, Msangi V, Tellevik MG, Maselle SY, et al. High rate of fatal cases of pediatric septicemia caused by gram-negative Bacteria with extended-Spectrum Beta-lactamases in Dar Es Salaam, Tanzania. J Clin Microbiol. 2005;43(2):745–749. doi: 10.1128/JCM.43.2.745-749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawat D, Nair D. Extended-spectrum β-lactamases in gram negative Bacteria. J Glob Infect Dis. 2010;2(3):263–274. doi: 10.4103/0974-777X.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruppé E, Woerther P, Barbier F. Mechanisms of antimicrobial resistance in gram-negative bacilli. Ann Intensive Care. 2015;5:21. doi: 10.1186/s13613-015-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagace-Wiens P, Nichol KA, Nicolle LE, Decorby RM, McCracken M, Alfa MJ, Mulvey MR, Zhanel GG. ESBL genotypes in fluoroquinolones-resistant and flouroquinolones-susceptible ESBL producing Escherichia coli urinary isolates in Manitoba. Can J Infect Dis Med Microbiol. 2007;2007(18):2. doi: 10.1155/2007/848194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henson SP, Boinett CJ, Ellington MJ, Kagia N, Mwarumba S, Nyongesa S, Mturi N, Kariuki S, Scott JAG, Thomson NR, et al. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int J Med Microbiol. 2017;307(7):422–429. doi: 10.1016/j.ijmm.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revathi G, Siu LK, Po-Liang L, Huang L. First report of NDM-1 producing Acinetobacter baumannii in East Africa. Int J Infect Dis. 2013;17:1255–1258. doi: 10.1016/j.ijid.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 29.Ntirenganya C, Manzi O, Muvunyi CM, Ogbuagu O. High prevalence of antimicrobial resistance among common bacterial isolates in a tertiary healthcare Facility in Rwanda. Am J Trop Med Hyg. 2015;92(4):865–870. doi: 10.4269/ajtmh.14-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ampaire LM, Katawera V, Nyehangane D, Boum Y, Bazira J. Epidemiology of Carbapenem resistance among multi-drug resistant Enterobacteriaceae in Uganda. Br Microbiol Res J. 2015;8(2):418–423. doi: 10.9734/BMRJ/2015/17055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kateete DP, Nakanjako R, Namugenyi J, Erume J, Joloba ML, Najjuka CF. Carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii at Mulago Hospital in Kampala, Uganda (2007–2009) Springerplus. 2016;5(1):1308. doi: 10.1186/s40064-016-2986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitout JDD, Revathid D, Chowa BL, Kariukie S, Nordmannf P, Poirel L. Metallo-β lactamase-producing Pseudomonas aeruginosa isolated from a large tertiary Centre in Kenya. Clin Microbiol. 2008;14(8):755–759. doi: 10.1111/j.1469-0691.2008.02030.x. [DOI] [PubMed] [Google Scholar]
- 33.Ssekatawa K, Byarugaba DK, Wampande E, Ejobi F. A systematic review: the current status of carbapenem resistance in East Africa. BMC Res Notes. 2018;11:629. doi: 10.1186/s13104-018-3738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desai K, Kazi M, Ajban K, Munshi M, Rodrigues C, Soman R, Shetty A. Clinical outcomes and safety of colistin in treatment of gram-negative infections: a prospective observational study. EJCCM. 2016;4(2):67–72. [Google Scholar]
- 35.Rousham EK, Unicomb L, Islam MA. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches. Proc Biol Sci. 2018;285(1876):20180332. doi: 10.1098/rspb.2018.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omulo S, Thumbi SM, Njenga MK, Call DR. A review of 40 years of enteric antimicrobial resistance research in eastern Africa: what can be done better? Antimicrob Resist Infect Control. 2015;4:1. doi: 10.1186/s13756-13014-10041-13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moyo SJ, Aboud S, Blomberg B, Mkopi N, Kasubi N, Manji K, Lyamuya EF, Maselle SY, Langeland N. High nasal carriage of Methicillin-resistant Staphylococcus aureus among healthy Tanzanian under-5 children. Microb Drug Resist. 2014;20:1. doi: 10.1089/mdr.2013.0016. [DOI] [PubMed] [Google Scholar]
- 38.Caudell MA, Mair C, Subbiah M, Matthews L, Quinlan RJ, Quinlan MB, Zadoks R, Keyyu J, Call DR. Identification of risk factors associated with carriage of resistant Escherichia coli in three culturally diverse ethnic groups in Tanzania: a biological and socioeconomic analysis. Lancet Planet Health. 2018;2:e489–e497. doi: 10.1016/S2542-5196(18)30225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleece M, Nshama R, Walongo T, Gratz J, Platts-Mills J, Mduma E, Houpt E. Risk Factors of Antibiotic Resistance in E. coli Isolated from the MAL-ED Birth Cohort Study in Rural Tanzania. Open Forum Infect Dis. 2018;5(1):S365–S366. [Google Scholar]
- 40.Joachim A, Moyo SJ, Nkinda L, Majigo M, Rugarabamu S, Mkashabani EG, Mmbaga EJ, Mbembati N, Aboud S, Lyamuya EF. Nasal carriage of methicillin-resistant Staphylococcus aureus among health care workers in tertiary and regional Hospitals in Dar es Salam, Tanzania. Int J Microbiol. 2018;2018:5058390. doi: 10.1155/2018/5058390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moyo S, Haldorsen B, Aboud S, Blomberg B, Maselle SY, Sundsfjord A, Langeland N, Ørjan Samuelsen Q. Identification of VIM-2-producing Pseudomonas aeruginosa from Tanzania is associated with sequence types 244 and 640 and the location of blaVIM-2 in a TniC Integron. Antimicrob Agents Chemother. 2015;59(1):682–685. doi: 10.1128/AAC.01436-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muvunyi CM, Masaisa F, Bayingana C, Mutesa L, Musemakweri A, Muhirwa G, Claeys GW. Decreased susceptibility to commonly used antimicrobial agents in bacterial pathogens isolated from urinary tract infections in Rwanda: need for new antimicrobial guidelines. Am J Trop Med Hyg. 2011;84(6):923–928. doi: 10.4269/ajtmh.2011.11-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kajeguka DC, Nambunga PP, Kabissi F, Kamugisha B, Kassam N, Nyombi B, Mataro C, Maro V, Chilongola JO. Antimicrobial resistance patterns of phenotype Extended Spectrum Beta-Lactamase producing bacterial isolates in a referral hospital in northern Tanzania. Tanzania J Health Res. 2015;17(3). 10.4314/thrb.v17i3.2.
- 44.Tellevik MG, Blomberg B, Kommedal Ø, Maselle SY, Langeland N, Moyo SJ: High Prevalence of Faecal Carriage of ESBL-Producing Enterobacteriaceae among Children in Dar es Salaam, Tanzania. PLoS One. 2016, 11(12): e0168024. 10.1371/journal.pone.0168024. [DOI] [PMC free article] [PubMed]
- 45.Mshana SE, Falgenhauer L, Mirambo MM, Mushi MF, Moremi N, Julius R, Seni J, Imirzalioglu C, Matee M, Chakraborty T. Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect Dis. 2016;16:187. doi: 10.1186/s12879-016-1527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moremi N, Claus H, Vogel U, Mshana SE. Faecal carriage of CTX-M extended-spectrum beta-lactamase-producing Enterobacteriaceae among street children dwelling in Mwanza city, Tanzania. PLoS One. 2017;12(9):e0184592. doi: 10.1371/journal.pone.0184592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaban SS. Prevalence of extended spectrum beta lactamase producing Escherichia coli in integrated agro-aquaculture in Morogoro, Tanzania. Morogoro: A dissertation submitted in partial fulfilment of the requirements for the degree of Master of Science in health of aquatic animal resources of Sokoine University of Agriculture; 2017. [Google Scholar]
- 48.Geofrey A, Abade A, Aboud S. Methicillin-resistant Staphylococcus aureus (MRSA) colonization among Intensive Care Unit (ICU) patients and health care workers at Muhimbili national hospital, Dar Es Salaam, Tanzania. Pan Afr Med J. 2015;21:211. doi: 10.11604/pamj.12015.11621.11211.14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basulira Y, Olet SA, Alele PE. Inappropriate usage of selected antimicrobials: Comparative residue proportions in rural and urban beef in Uganda. PLoS One. 2019;14(1):e0209006. doi: 10.1371/journal.pone.0209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marando R, Seni J, Mirambo MM, Falgenhauer L, Moremi N, Mushi MF, Kayangea NM, Manyama F, Imirzalioglu C, Chakraborty T, et al. Predictors of the extended-spectrum-beta lactamases producing Enterobacteriaceae neonatal sepsis at a tertiary hospital, Tanzania. Int J Med Microbiol. 2018;308(7):803–811. doi: 10.1016/j.ijmm.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butaye P, Devriese LA, Freddy Haesebrouck F. Antimicrobial growth promoters used in animal feed: effects of less well-known antibiotics on gram-positive Bacteria. Clin Microbiol Rev. 2003;16(2):175–188. doi: 10.1128/CMR.16.2.175-188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 2011;24:718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep. 2012;127:4–22. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asokan GV, Kasimanickam RK. Emerging infectious diseases, antimicrobial resistance and millennium development goals: resolving the challenges through one health. Central Asian J Glob Health. 2013;2:2. doi: 10.5195/cajgh.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sonda T, Kumburu H, van Zwetselaar M, Alifrangis M, Mmbaga BT, Lund O, Aarestrup FM, Kibiki G. Prevalence and risk factors for CTX-M gram-negative bacteria in hospitalized patients at a tertiary care hospital in Kilimanjaro, Tanzania. Eur J Clin Microbiol Infect Dis. 2018;37(5):897–906. doi: 10.1007/s10096-018-3196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cotton MF, Wasserman E, Smit J, Whitelaw A, Zar HJ. High incidence of antimicrobial resistant organisms including extended spectrum beta-lactamase producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus in nasopharyngeal and blood isolates of HIV-infected children from Cape Town, South Africa. BMC Infect Dis. 2008;8:40. doi: 10.1186/1471-2334-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rameshkumar MR, Arunagirinathan A. Drug-resistant bacterial infections in HIV chapter in advances in HIV and AIDS control, patients. 2018. [Google Scholar]
- 58.Kolár M, Urbánek K, Látal T. Antibiotic selective pressure and development of bacterial resistance. Int J Antimicrob Agents. 2001;17(5):357–363. doi: 10.1016/s0924-8579(01)00317-x. [DOI] [PubMed] [Google Scholar]
- 59.Cantón R, Morosini M-I. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev. 2011;35(5):977–991. doi: 10.1111/j.1574-6976.2011.00295.x. [DOI] [PubMed] [Google Scholar]
- 60.Knöppel A, Näsvall J, Andersson DI. Evolution of antibiotic resistance without antibiotic exposure. Evolution of antibiotic resistance without antibiotic exposure. Antimicrob Agents Chemother. 2017;61:e01495–e01417. doi: 10.1128/AAC.01495-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isendahl J, Turlej-Rogacka A, Manjuba C, Rodrigues A, Giske CG, Nauclér P. Fecal carriage of ESBL-producing E. coli and K. pneumoniae in children in Guinea-Bissau: a hospital-based cross-sectional study. PLoS One. 2012;2012(7):e51981. doi: 10.1371/journal.pone.0051981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lonchel CM, Melin P, Gangoué-Piéboji J, Assoumou M-CO, Boreux R, De Mol P. Extended-spectrum β-lactamase-producing Enterobacteriaceae in Cameroonian hospitals. Eur J Clin Microbiol Infect Dis. 2013;32(1):79–87. doi: 10.1007/s10096-012-1717-4. [DOI] [PubMed] [Google Scholar]
- 63.Moremi N, Claus H, Rutta L, Frosch M, Vogel U, Mshana SE. High carriage rate of extended-spectrum beta-lactamase-producing Enterobacteriaceae among patients admitted for surgery in Tanzanian hospitals with a low rate of endogenous surgical site infections. J Hosp Infect. 2018;100(1):47–53. doi: 10.1016/j.jhin.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 64.Seni J, Tito JN, Makoye SJ, Mbena H, Alfred HS, van der Meer F, Pitout JDD, Mshana SE, DeVinney R. Multicentre evaluation of significant bacteriuria among pregnant women in the cascade of referral healthcare system in North-Western Tanzania: bacterial pathogens, antimicrobial resistance profiles and predictors. J Glob Antimicrob Resist. 2019;17:173–179. doi: 10.1016/j.jgar.2018.12.024. [DOI] [PubMed] [Google Scholar]
- 65.Seni J, Mwakyoma AA, Mashuda F, Marando R, Ahmed M, DeVinney R, Pitout JDD, Mshana SE. Deciphering risk factors for blood stream infections, bacteria species and antimicrobial resistance profiles among children under five years of age in North-Western Tanzania: a multicentre study in a cascade of referral health care system. BMC Pediatr. 2019;19:32. doi: 10.1186/s12887-019-1411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moremi N, Claus H, Vogel U, Mshana SE. The role of patients and healthcare workers Staphylococcus aureus nasal colonization in occurance of surgical site infection among patients admitted in two centers in Tanzania. Antimicrob Resist Infect Control. 2019;8:102. doi: 10.1186/s13756-019-0554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Armah EO. A dissertation submitted in partial fulfilment of the requirements for the degree of Master of Science in molecular biology and biotechnology of Sokoine University of Agriculture, Morogoro, Tanzania. 2016. [Google Scholar]
- 68.Albrechtova K, Dolejska M, Cizek A. D T, Klimes J, Bebora L, Literaka I: dogs of nomadic pastoralists in northern Kenya are reservoirs of plasmid-mediated cephalosporin- and quinolone-resistant Escherichia coli, including pandemic clone B2-O25-ST131. Antimicrob Agents Chemother. 2012;56(7):4013–4017. doi: 10.1128/AAC.05859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moremi N, Manda EV, Falgenhauer L, Ghosh H, Imirzalioglu C, Matee M, Chakraborty T, Mshana SE. Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front Microbiol. 2016;7:1862. doi: 10.3389/fmicb.2016.01862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mshana SE, Imirzalioglu C, Hain T, Domann E, Lyamuya EF, Chakraborty T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect. 2011;17(8):1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x. [DOI] [PubMed] [Google Scholar]
- 71.Kumburu HH, Sonda T, Leekitcharoenphon P, van Zwetselaar M, Lukjancenko O, Alifrangis M, Lund O, Mmbaga BT, Kibiki G, Aarestrup FM. Hospital Epidemiology of Methicillin-Resistant Staphylococcus aureus in a Tertiary Care Hospital in Moshi, Tanzania, as Determined by Whole Genome Sequencing. BioMed Res Int. 2018;2018:2087693. doi: 10.1155/2018/2087693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maina EK, Kiiyukia C, Njeri Wamae C, Waiyaki PG, Kariuki S. Characterization of methicillin-resistant Staphylococcus aureus from skin and soft tissue infections in patients in Nairobi, Kenya. Int J Infect Dis. 2013;17(2):e115–e119. doi: 10.1016/j.ijid.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Kateete DP, Namazzi S, Okee M, Okeng A, Baluku H, Musisi NL, Katabazi FA, Joloba ML, Ssentongo R, Najjuka FC. High prevalence of methicillin resistant Staphylococcus aureus in the surgical units of Mulago hospital in Kampala, Uganda. BMC Res Notes. 2011;4:326. doi: 10.1186/1756-0500-4-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moremi N, Mshana SE, Kamugisha E, Kataraihya JB, Tappe D, Vogel U, Lyamuya EF, Claus F. Predominance of methicillin resistant Staphylococcus aureus-ST88 and new ST1797 causing wound infection and abscesses. J Infect Dev Ctries. 2012;6(8):620–625. doi: 10.3855/jidc.2093. [DOI] [PubMed] [Google Scholar]
- 75.Omuse G, Van Zyl KN, Hoek K, Abdulgader S, Kariuki S, Whitelaw A, Revathi G. Molecular characterization of Staphylococcus aureus isolates from various healthcare institutions in Nairobi, Kenya: a cross sectional study. Ann Clin Microbiol Antimicrob. 2016;15:51. doi: 10.1186/s12941-016-0171-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seni J, Bwanga F, Najjuka CF, Makobore P, Okee M, Mshana SE, Kidenya BR, Joloba ML, Kateete DP. Molecular Characterization of Staphylococcus aureus from Patients with Surgical Si Infections at Mulago Hospital in Kampala, Uganda. PLoS One. 2013;8(6):e66153. doi: 10.1371/journal.pone.0066153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aiken AM, Mutuku IM, Sabat AJ, Akkerboom V, Mwangi J, Scott JAG, Morpeth SC, Friedrich AW, Grundmann H. Carriage of Staphylococcus aureus in Thika level 5 hospital, Kenya: a cross-sectional study. Antimicrob Resist Infect Control. 2014;3:22. doi: 10.1186/2047-2994-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Asiimwe BB, Baldan R, Trovato A, Cirillo DM. Prevalence and molecular characteristics of Staphylococcus aureus, including methicillin resistant strains, isolated from bulk can milk and raw milk products in pastoral communities of south-West Uganda. BMC Infect Dis. 2017;17:422. doi: 10.1186/s12879-017-2524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anjum MF, Lemma F, Cork DJ, Meunier D, Murphy N, North SE, Woodford N, Haines J, Randall LP. (Isolation and detection of extended spectrum β-lactamas ESBL)-producing Enterobacteriaceae from meat using chromogenic agars and isothermal loop-mediated amplification (LAMP) assays. J Food Sci. 2013;78:1892–1898. doi: 10.1111/1750-3841.12297. [DOI] [PubMed] [Google Scholar]
- 80.Poirel L, Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front Microbiol. 2012;3:1–7. doi: 10.3389/fmicb.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moremi N, Claus H, Silago V, Kabage K, Abednego R, Matee M, Vogel U, Mshana SE. Hospital surface contamination with antimicrobial-resistant gram-negative organisms in Tanzanian regional and tertiary hospitals: the need to improve environmental cleaning. J Hosp Infect. 2019;102(1):98–100. doi: 10.1016/j.jhin.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Tekiner IH, Özpınar H. Occurrence and characteristics of extended spectrum beta-lactamases-producing Enterobacteriaceae from foods of animal origin. Braz J Microbiol. 2016;47(2):444–451. doi: 10.1016/j.bjm.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khasabuli OY, Ngugi C, Kiiru J. Carriage, antimicrobial susceptibility profiles and genetic diversity of Staphylococcus aureus and MRSA isolates recovered from students in a Kenyan university. East Afr Med J. 2017;94:7. [Google Scholar]
- 84.Couce A, Rodríguez-Rojas A, Blázquez J. Determinants of genetic diversity of spontaneous drug resistance in Bacteria. Genetics. 2016;203(3):1369–1380. doi: 10.1534/genetics.115.185355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paterson DL, Bonomo RA. Extended-Spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moyo SJ, Aboud S, Kasubi M, Maselle SY. Bacterial isolates and drug susceptibility patterns of urinary tract infection among pregnant women at Muhimbili National Hospital in Tanzania. Tanzania J Health Res. 2010;12(4):236–240. doi: 10.4314/thrb.v12i4.52997. [DOI] [PubMed] [Google Scholar]
- 87.Wangai FK, Masika MM, Maritim MC, Seaton RA. Methicillin-resistant Staphylococcus aureus (MRSA) in East Africa: red alert or red herring? BMC Infect Dis. 2019;19:596. doi: 10.1186/s12879-019-4245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gitau W, Masika M, Musyoki M, Museve B, Titus Mutwiri T. Antimicrobial susceptibility pattern of Staphylococcus aureus isolates from clinical specimens at Kenyatta National Hospital. BMC Res Notes. 2018;11:226. doi: 10.1186/s13104-018-3337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matee MI. Antimicrobial resistance (AMR) at the Southern African Centre for Infectious Diseases Surveillance (SACIDS) 2019. [Google Scholar]
- 90.Kairuki S, Keddy HK, Antonio M. Antimicrobial resistance surveillance in Africa: successes, gaps and a roadmap for the future. Afr J Lab Med. 2018;7(2):924. doi: 10.4102/ajlm.v7i2.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guerra-Assunção JA, Crampin AC, Houben RM, Mzembe T, Mallard K, Coll F, Khan P, Banda L, Chiwaya A, Pereira RP, et al. Large-scale whole genome sequencing of M tuberculosis provides insights into transmission in a high prevalence area. Elife. 2015;3:4. doi: 10.7554/eLife.05166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Phelan JE, O’Sullivan DM, Machado D, Ramos J, Oppong YEA, Campino S, O’Grady J, McNerney R, Hibberd ML, Viveiros M, Huggett JF, Clark TG. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 2019;11:41. doi: 10.1186/s13073-019-0650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data and materials were available from peer reviewed articles published between January 2001 and December 2018 in East Africa.