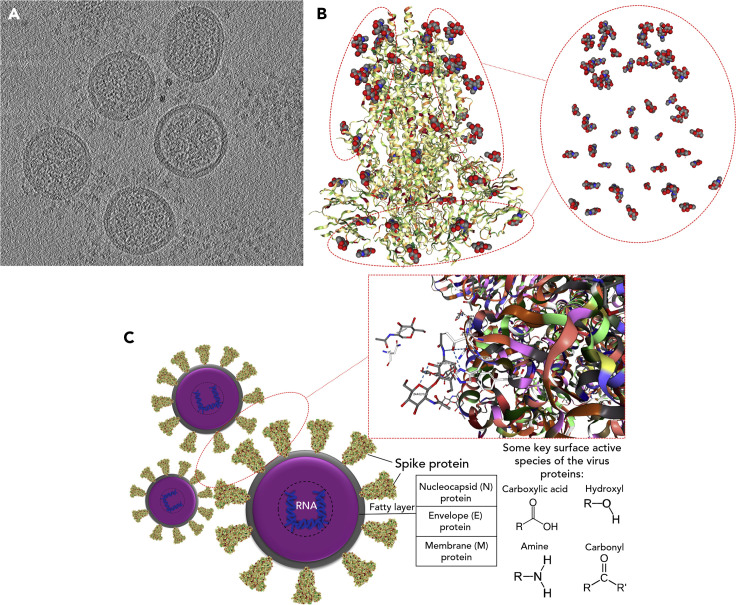
Figure 1.
Structural Chemistry and Identifying Surface-Active Species of the Virus
(A) A central slice through a cryo-EM tomogram of mouse hepatitis virus for showing the existence of S on the outer surface of virions.
(B) Structure of the SARS-CoV-2 spike glycoprotein with highlighting corresponding functional groups (protein database [PDB]: 6VXX). Grey, blue, and red spheres are carbon, nitrogen, and oxygen atoms, respectively. The molecular structure is colored based on hydrophobicity; colored from red (hydrophilic) to green (hydrophobic). It utilizes the experimentally attained hydrophobicity scale that relied on whole-residue free energies of transfer ΔG (kcal/mol) from water to 1-Palmitoyl-2-oleoylphosphatidylcholine (POPC) interface.
(C) Model representation of SARS-CoV-2 with respective proteins assembly. Key molecular interactions among proteins on the surface of the virus particle are shown as gray dash lines, “hydrophobic interactions,” and blue dash lines, “hydrogen bonding (–O–H···O),” (PDB: 6VYB). Some key surface-active moieties of SARS-CoV-2 are denoted as hydroxyl, amine, carbonyl, and carboxylic acid functional groups.