Here, we report the draft genome sequences of six severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strains. SARS-CoV-2 is responsible for the COVID-19 pandemic, which started at the end of 2019 in Wuhan, China. The isolates were obtained from nasopharyngeal swabs from Moroccan patients with COVID-19. Mutation analysis revealed the presence of the spike D614G mutation in all six genomes, which is widely present in several genomes around the world.
ABSTRACT
Here, we report the draft genome sequences of six severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strains. SARS-CoV-2 is responsible for the COVID-19 pandemic, which started at the end of 2019 in Wuhan, China. The isolates were obtained from nasopharyngeal swabs from Moroccan patients with COVID-19. Mutation analysis revealed the presence of the spike D614G mutation in all six genomes, which is widely present in several genomes around the world.
ANNOUNCEMENT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is classified within the subgenus Sarbecovirus and genus Betacoronavirus and was first identified in Wuhan, China (1), as the causative agent for COVID-19 disease. Since then, the number of COVID-19 cases has risen dramatically (2).
In Morocco, the first SARS-CoV-2 case was confirmed on 2 March 2020. As of 29 June 2020, the number of cases had reached more than 12,248. To understand SARS-CoV-2 genetic diversity and molecular epidemiology in Morocco, we performed complete genome sequencing using the Oxford Nanopore MinION technology.
In this study, we announce the genome sequences of six SARS-CoV-2 strains isolated from patients in Morocco. The samples were obtained by taking nasopharyngeal swabs from six patients with COVID-19. The viral RNA was extracted directly from the swab using the QIAamp viral RNA minikit (Qiagen, Germany), and the Transcriptor first-strand cDNA synthesis kit (Roche) with random hexamers was used to synthetize the viral cDNA.
The ARTIC v3 primers were used with the Q5 high-fidelity DNA polymerase (New England BioLabs [NEB], USA) for virus DNA enrichment. Amplicons of 400 bp were purified using sample purification beads (SPBs) (Illumina, USA) (3) and then quantified with a Qubit 3.0 fluorometer and used for library preparation.
Sequencing was performed on a MinION MK1C instrument with a ligation sequencing kit (catalog number SQK-LSK109) according to a standard protocol (Oxford Nanopore Technologies [ONT], UK), and the six samples were multiplexed in one run. The R9 flow cell was used and run for 2 h.
The sequence reads generated were between 70,565 and 185,364 (Table 1) of raw data per sample, with average lengths of 454 bp, 455 bp, 455 bp, 452 bp, 455 bp, and 454 bp for strains RMPS-01, RMPS-02, RMPS-03, RMPS-04, RMPS-05, and RMPS-06, respectively. Raw reads were mapped to a SARS-CoV-2 reference genome under GenBank accession number NC_045512.2 using BWA-MEM v. 0.7.17 for single-end reads with default settings (4), and SAM/BAM files were manipulated by SAMtools v. 1.9-11 (5). Variant calling was performed using BCFtools v. 1.9 with “mpileup” (5), and variants were further annotated using SnpEff v. 4.3T (6). The consensus sequences were generated by mapping the variants to the reference genomes using BCFtools (5) and then were submitted to the GISAID database and NCBI (accession numbers are listed in Table 1).
TABLE 1.
Genome features of six strains of SARS-CoV-2
| Strain | GenBank accession no. | NCBI SRA accession no. | No. of raw reads | Genome size (bp) | GC content (%) | Coverage (×) | Mapped read %a |
|---|---|---|---|---|---|---|---|
| RMPS-01 | MT731285 | SRX8633309 | 71,570 | 29,903 | 37.96 | 870.2 | 99.93 |
| RMPS-02 | MT731292 | SRX8633310 | 185,364 | 29,903 | 37.96 | 2,213.9 | 98.92 |
| RMPS-03 | MT731673 | SRX8633311 | 88,452 | 29,903 | 37.96 | 1,022.5 | 96.51 |
| RMPS-04 | MT731327 | SRX8633312 | 113,813 | 29,903 | 37.96 | 1,291 | 94.9 |
| RMPS-05 | MT731468 | SRX8633313 | 70,565 | 29,903 | 37.96 | 834.5 | 98.2 |
| RMPS-06 | MT731764 | SRX8633314 | 128,615 | 29,903 | 37.96 | 1,291 | 94.9 |
Refers to the percentage of reads from the sequenced sample that align directly to a single region on the reference genome.
The phylogenetic analysis was realized using 250 genome sequences retrieved from the GISAID database. The alignment was performed using MAFFT (7) for fast alignment, and maximum-likelihood trees were inferred with IQ-TREE v. 1.5.5 under the generalized time-reversable (GTR) model (8), implemented via the pipeline provided by Augur (github.com/nextstrain/augur). The generated tree was visualized using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree). Major clades were defined by amino acid and/or nucleotide substitutions and were matched to the Nextstrain nomenclature (9) (https://nextstrain.org/ncov).
The size of the consensus sequences was similar to that of the Wuhan-Hu-1 reference genome (GenBank accession number NC_045512.2) and was 29,903 bp with a mean coverage ranging from 843.5× to 2,213×. The strain details are found in Table 1.
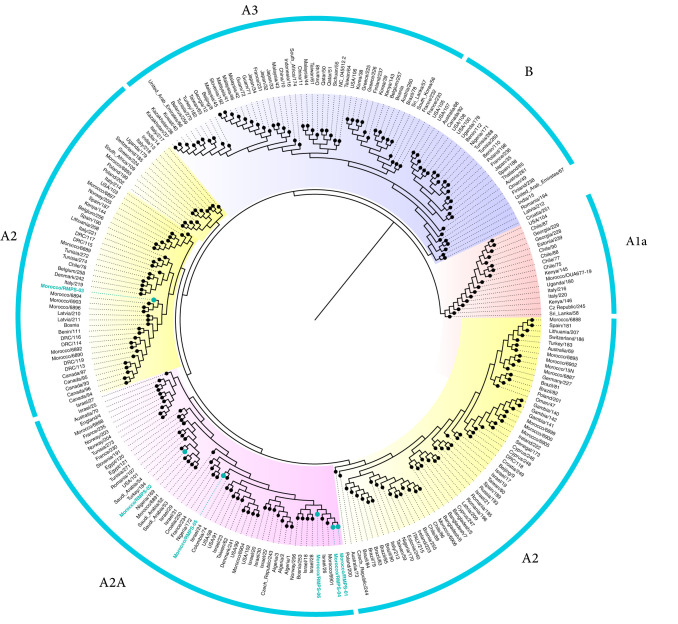
We detected 16 different variants in the 6 analyzed genomes. All the genomes shared four mutations, namely, two synonymous (F924F and L4715L), one nonsynonymous (D614G), and one intergenic (241C>T). Only one nonsynonymous mutation was detected (D614G) in the spike protein, which is known as the most prevalent variant worldwide (10), and it is also associated with the emergence of clade A2, which includes all Moroccan strains sequenced in this study (Fig. 1). This mutation was already associated with the observed transmission increase in the United States (10–12).
FIG 1.
Phylogenetic tree of six SARS-CoV-2 genomes from Morocco. We are currently sequencing more genomes from Morocco to further investigate the spread of COVID-19 and to monitor the evolution of SARS-CoV-2 in Morocco.
Data availability.
The reads of the six SARS-CoV-2 strains were deposited in DDBJ/ENA/GenBank under the SRA accession numbers SRR12109250, SRR12109251, SRR12109252, SRR12109253, SRR12109254, and SRR12109255. The consensus sequences were also deposited in GenBank under the accession numbers MT731285, MT731292, MT731673, MT731327, MT731468, and MT731764.
ACKNOWLEDGMENTS
This work was carried out under national funding from the Moroccan Ministry of Higher Education and Scientific Research (COVID-19 program) to A.I. This work was also supported by a grant from the Moroccan Institute of Cancer Research and the PPR-1 program to A.I.
We declare no competing interests.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan T. 2020. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evol 81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.ARTIC. 2020. nCoV-2019 sequencing protocol. https://www.protocols.io/view/ncov-2019-sequencing-protocol-bbmuik6w.
- 4.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Group . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Jackson CB, Mou H, Ojha A, Rangarajan ES, Izard T, Farzan M, Choe H. 2020. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv doi: 10.1101/2020.06.12.148726. [DOI]
- 11.Alouane T, Laamarti M, Essabbar A, Hakmi M, Bouricha EM, Chemao-Elfihri MW, Kartti S, Boumajdi N, Bendani H, Laamrti R, Ghrifi F, Allam L, Aanniz T, Ouadghiri M, El Hafidi N, El Jaoudi R, Benrahma H, Elattar J, Mentag R, Sbabou L, Nejjari C, Amzazi S, Belyamani L, Ibrahimi A. 2020. Genomic diversity and hotspot mutations in 30,983 SARS-CoV-2 genomes: moving toward a universal vaccine for the “confined virus”? bioRxiv doi: 10.1101/2020.06.20.163188. [DOI] [PMC free article] [PubMed]
- 12.Laamarti M, Alouane T, Kartti S, Chemao-Elfihri MW, Hakmi M, Essabbar A, Laamart M, Hlali H, Allam L, Hafidi NEL, Jaoudi REL, Allali I, Marchoudi N, Fekkak J, Benrahma H, Nejjari C, Amzazi S, Belyamani L, Ibrahimi A. 2020. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geo-distribution and a rich genetic variations of hotspots mutations. bioRxiv doi: 10.1101/2020.05.03.074567. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The reads of the six SARS-CoV-2 strains were deposited in DDBJ/ENA/GenBank under the SRA accession numbers SRR12109250, SRR12109251, SRR12109252, SRR12109253, SRR12109254, and SRR12109255. The consensus sequences were also deposited in GenBank under the accession numbers MT731285, MT731292, MT731673, MT731327, MT731468, and MT731764.