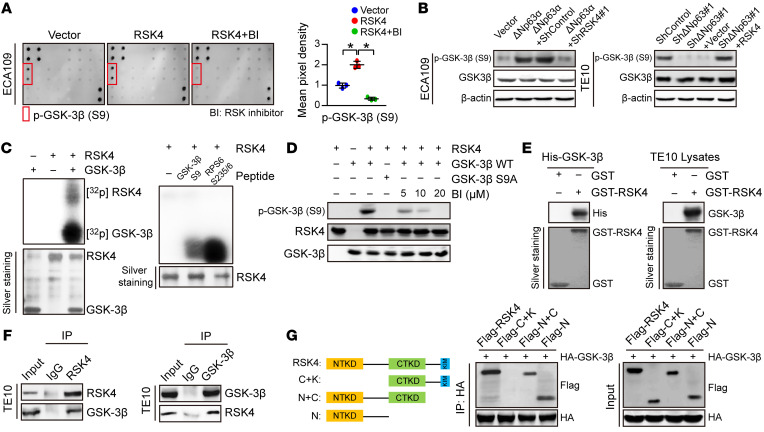
Figure 6. RSK4 directly phosphorylates GSK-3β (Ser9).
(A) MAPK pathway phosphorylated antibody microarray analysis shows that p–GSK-3β (Ser9) was significantly increased when RSK4 was stably overexpressed and decreased when treated with 10 μM BI-D1870 (an inhibitor of RSK) for 12 hours (n = 3 independent experiments). (B) Western blot analysis showing that downregulation of RSK4 resulted in a reduced level of ΔNp63α-induced p–GSK-3β (Ser9) (S9) (left), whereas RSK4 overexpression partially reversed the reduction in phosphorylation levels through ΔNp63 knockdown (right). (C) Active RSK4 phosphorylated GSK-3β at Ser9 in vitro in the presence of [γ-32P] ATP as visualized by an autoradiograph. The RPS6 (Ser235/236) peptide was used as a positive control. The input was confirmed by silver staining. (D) Validation of p–GSK-3β (Ser9) phosphorylation levels in an in vitro kinase assay by Western blotting. The levels of p–GSK-3β were inhibited when treated with BI-D1870 for 2 hours. WT GSK-3β and mutant GSK-3β S9A were used as substrates for active RSK4. (E) An in vitro GST-pulldown assay was performed to verify the interaction of RSK4 with purified His–GSK-3β protein (left) or GSK-3β from TE10 cell lysates (right). Retrieved proteins were evaluated by immunoblotting. GST-only protein was used as a negative control. GST fusion proteins were confirmed by silver staining. (F) The interaction of RSK4 and GSK-3β was confirmed by an endogenous co-IP assay in TE10 cells. IgG served as a negative control. (G) Mapping analyses of full-length and truncated RSK4 with representative co-IP assays in HEK293T cells showing that the NTKD of RSK4 was responsible for the interaction with GSK-3β. C, CTKD; K, kinase interaction motif (KIM); N, NTKD. Data represent the mean ± SD. *P < 0.05. Differences were tested using 1-way ANOVA with Tukey’s post hoc test (A).