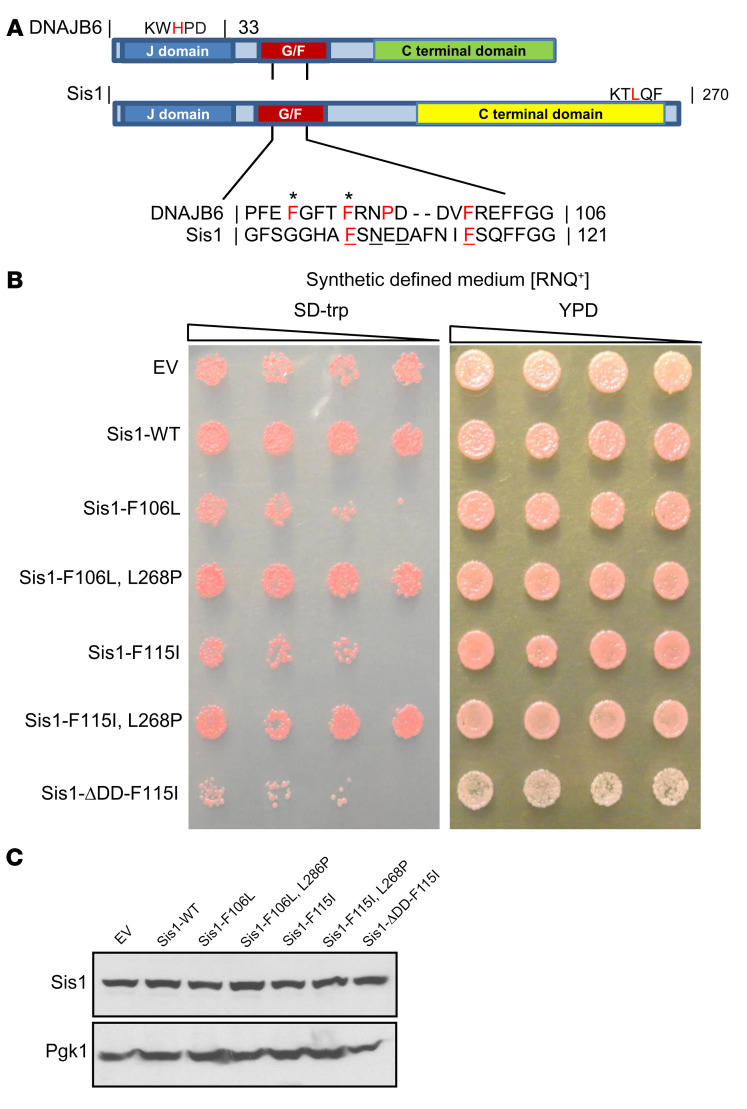
Figure 1. LGMDD1 mutations are toxic in yeast Sis1 and rescued by impairing HSP70 association.
(A) Domain alignment of DNAJB6 and Sis1. Amino acid alignment of the G/F domains denotes sequence homology. Amino acids in bold red represent the mutations used in the study. Amino acids in red represent other disease mutations. Underlined amino acids represent DNAJB6 and Sis1 homologous residues mutated in LGMDD1. The amino acids with asterisks represent residues mutated in our 2 animal models. (B) Yeast cells propagating single-dot medium [RNQ+] were transformed with constructs overexpressing Sis1-WT, Sis1-F106L (DNAJB6-F93L homologous), Sis1-F115I (DNAJB6-F100I homologous), or empty vector control (EV). Yeast were also transformed with Sis1-mutant constructs containing a second mutation that impairs HSP70 association (L268P) or blocks Sis1 dimerization (ΔDD). Cultures were normalized and serially diluted 5-fold and were spotted on medium (SD-trp) to select for the plasmid or medium (yeast extract peptone dextrose [YPD]) that provided no selection. (C) Western blot analysis from lysates of yeast expressing the Sis1 constructs in B. Pgk1 was used as a loading control. Images in A and B are representative of 3 independent experiments.